Synthetic genomes unveil the effects of synonymous recoding
Synthetic genomes unveil the effects of synonymous recoding
Nyerges, A.; Chiappino-Pepe, A.; Budnik, B.; Baas-Thomas, M.; Flynn, R.; Yan, S.; Ostrov, N.; Liu, M.; Wang, M.; Zheng, Q.; Hu, F.; Chen, K.; Rudolph, A.; Chen, D.; Ahn, J.; Spencer, O.; Ayalavarapu, V.; Tarver, A.; Harmon-Smith, M.; Hamilton, M.; Blaby, I.; Yoshikuni, Y.; Hajian, B.; Jin, A.; Kintses, B.; Szamel, M.; Seregi, V.; Shen, Y.; Li, Z.; Church, G. M.
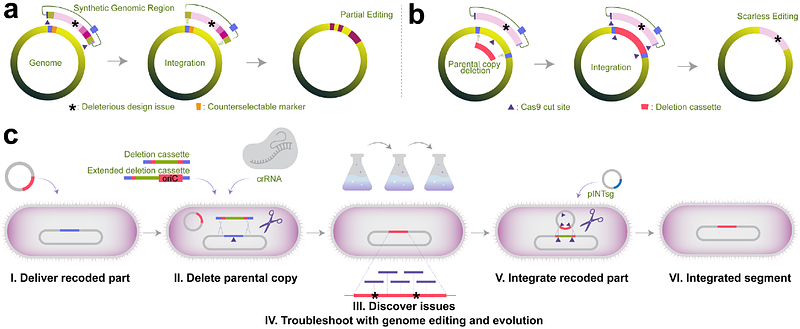
AbstractEngineering the genetic code of an organism provides the basis for (i) making any organism safely resistant to natural viruses and (ii) preventing genetic information flow into and out of genetically modified organisms while (iii) allowing the biosynthesis of genetically encoded unnatural polymers. Achieving these three goals requires the reassignment of multiple of the 64 codons nature uses to encode proteins. However, synonymous codon replacement--recoding--is frequently lethal, and how recoding impacts fitness remains poorly explored. Here, we explore these effects using whole-genome synthesis, multiplexed directed evolution, and genome-transcriptome-translatome-proteome co-profiling on multiple recoded genomes. Using this information, we assemble a synthetic Escherichia coli genome in seven sections using only 57 codons to encode proteins. By discovering the rules responsible for the lethality of synonymous recoding and developing a data-driven multi-omics-based genome construction workflow that troubleshoots synthetic genomes, we overcome the lethal effects of 62,007 synonymous codon swaps and 11,108 additional genomic edits. We show that synonymous recoding induces transcriptional noise including new antisense RNAs, leading to drastic transcriptome and proteome perturbation. As the elimination of select codons from an organism\'s genetic code results in the widespread appearance of cryptic promoters, we show that synonymous codon choice may naturally evolve to minimize transcriptional noise. Our work provides the first genome-scale description of how synonymous codon changes influence organismal fitness and paves the way for the construction of functional genomes that provide genetic firewalls from natural ecosystems and safely produce biopolymers, drugs, and enzymes with an expanded chemistry.


