Dynamical Responses Predict a Distal Site that Modulates Activity in an Antibiotic Resistance Enzyme
Dynamical Responses Predict a Distal Site that Modulates Activity in an Antibiotic Resistance Enzyme
Beer, M.; Oliveira, A. S. F.; Tooke, C. L.; Hinchliffe, P.; Li, A. T. Y.; Balega, B.; Spencer, J.; Mulholland, A. J.
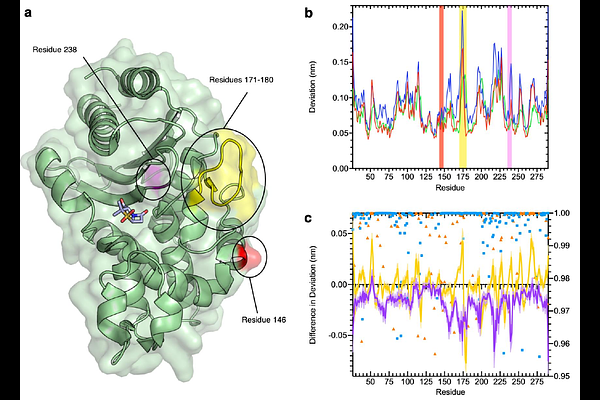
Abstract{beta}-lactamases, which hydrolyse {beta}-lactam antibiotics, are key determinants of antibiotic resistance. Predicting the sites and effects of distal mutations in enzymes is challenging. For {beta}-lactamases, the ability to make such predictions would contribute to understanding activity against, and development of, antibiotics and inhibitors to combat resistance. Here, using dynamical non-equilibrium molecular dynamics (D-NEMD) simulations combined with experiments, we demonstrate that intramolecular communication networks differ in three class A SulpHydryl Variant (SHV)-type {beta}-lactamases). Differences in network architecture and correlated motions link to catalytic efficiency and {beta}-lactam substrate spectrum. Further, the simulations identify a distal residue 89 in the clinically important Klebsiella pneumoniae carbapenemase 2 (KPC-2), as a participant in similar networks, suggesting that mutation at this position would modulate enzyme activity. Experimental kinetics, biophysical and structural characterisation of the naturally occurring, but previously biochemically uncharacterised, KPC-2 G89D mutant with several antibiotics and inhibitors reveals significant changes in hydrolytic spectrum, specifically reducing activity towards carbapenems without effecting major structural or stability changes. These results show that D-NEMD simulations can predict distal sites where mutation affects enzyme activity. This approach could have broad application in understanding enzyme evolution, and in engineering of natural and de novo enzymes.