KODAMA enables self-guided weakly supervised learning in spatial transcriptomics
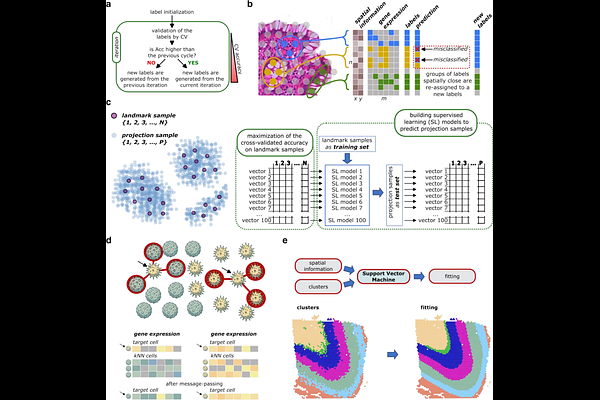
KODAMA enables self-guided weakly supervised learning in spatial transcriptomics
Abdel-Shafy, E. A.; Kassim, M.; Vignoli, A.; Mamdouh, F.; Tyekucheva, S.; Ahmed, D.; Ojo, D.; Price, B.; Socciarelli, F.; Duarte-Delgado, N. P.; Ocharo, M.; Okeke, C. J.; Triboli, L.; MacIntyre, D. A.; Loda, M.; Sarkar, D.; Gupta, D.; Piazza, S.; Zerbini, L. F.; Tenori, L.; Cacciatore, S.
AbstractSpatial transcriptomics provides researchers with a powerful tool to investigate gene expression patterns within their native positional context in tissues, revealing intricate cellular relationships. However, analyzing spatial transcriptomics data presents unique challenges due to their high dimensionality and complexity. Several competitive tools have emerged, each aiming to integrate spatial dependencies into their workflow. Yet, the overall performance of these approaches remains constrained by limitations in prediction accuracy and compatibility with specific data types. Here, we introduce the third version of the KODAMA algorithm, specifically tailored for spatial transcriptomics analysis. This upgraded version incorporates a novel approach that effectively reduces data dimensionality while preserving spatial information. At its core, the KODAMA algorithm employs parallel iterations that enforce spatial constraints throughout an embedded clustering process. Our method provides the flexibility to simultaneously analyze multiple samples, streamlining analysis workflows. Additionally, it extends beyond traditional 2D analysis to accommodate multidimensional datasets, including 3D spatial information. KODAMA seamlessly integrates into various pipelines, such as Seurat and Giotto. Extensive evaluations of KODAMA on 10x Visium, Visium HD, and image-based 3D datasets demonstrate its high accuracy in spatial domain prediction. Comparative analyses against alternative dimensionality reduction techniques and spatial analysis tools consistently validate and highlight KODAMA\'s superior performance in unraveling the spatial organization of cellular components across both single and integrated tissue samples.