Leveraging Dynamic Stability to Infer Regulation in Protein-Protein Interaction Networks: A Study of Infectious Vulnerability in COPD.
Leveraging Dynamic Stability to Infer Regulation in Protein-Protein Interaction Networks: A Study of Infectious Vulnerability in COPD.
Reimer, J.; Page, J.; Saha, P.; Shen, S.; Zhu, X.; Qian, S.; Mammen, M.; Qu, J.; Sethi, S.; Broderick, G. J.
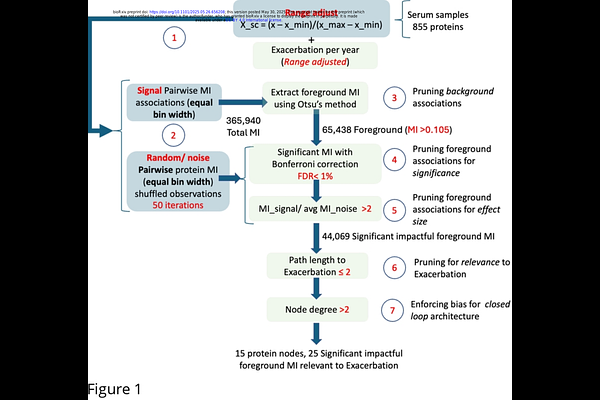
AbstractThe fourth leading cause of death in the US, Chronic Obstructive Pulmonary Disease (COPD) is punctuated by frequent viral and bacterial infections causing severe acute exacerbations (AECOPD) and increased mortality. In previous work we have shown that altered immune cell signaling may confer increased and persistent susceptibility to infection. Here we continue this investigation by conducting broad-spectrum proteomic profiling of circulating white blood cells to assemble an empirical protein-protein interaction network associated with frequency of infectious exacerbation. In a novel extension of conventional cross-sectional data analyses, we translate these undirected protein-protein interactions into candidate regulatory relationships with both direction and mode of action. The latter are inferred by formulating and solving a constraint satisfaction problem (SAT) whereby predicted dynamic behaviors of any valid regulatory network must support the expected persistent nature of low and high vulnerability phenotypes. Solving this SAT problem produced a set of competing candidate protein regulatory network architectures and signalling rules that unanimously highlighted several novel candidate pathway elements involved in oxidative stress response. Analysis of the overall dynamics supported by these networks, again supported the hypothesis that progression beyond an immune tipping point may confer persistent susceptibility to infection and that this may constitute a stable phenotype or regulatory trap in COPD characterized by a reactive oxygen cascade.