Reverse engineering neuron type-specific and type-orthogonal splicing-regulatory networks using single-cell transcriptomes
Reverse engineering neuron type-specific and type-orthogonal splicing-regulatory networks using single-cell transcriptomes
Moakley, D. F.; Campbell, M.; Anglada-Girotto, M.; Feng, H.; Califano, A.; Au, E.; Zhang, C.
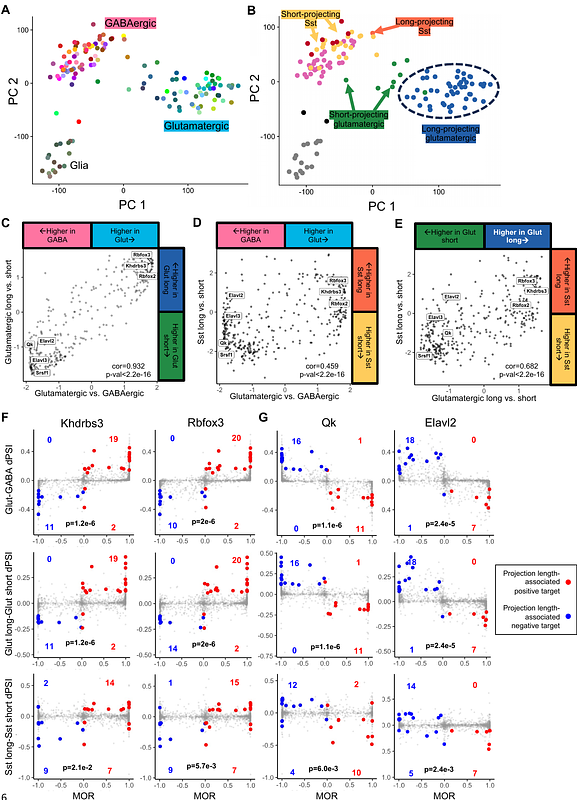
AbstractCell type-specific alternative splicing (AS) enables differential gene isoform expression between diverse neuron types with distinct identities and functions. Current studies linking individual RNA-binding proteins (RBPs) to AS in a few neuron types underscore the need for holistic modeling. Here, we use network reverse engineering to derive a map of the neuron type-specific AS regulatory landscape from 133 mouse neocortical cell types defined by single-cell transcriptomes. This approach reliably inferred the regulons of 350 RBPs and their cell type-specific activities. Our analysis revealed driving factors delineating neuronal identities, among which we validated Elavl2 as a key RBP for MGE-specific splicing in GABAergic interneurons using an in vitro ESC differentiation system. We also identified a module of exons and candidate regulators specific for long- and short-projection neurons across multiple neuronal classes. This study provides a resource for elucidating splicing regulatory programs that drive neuronal molecular diversity, including those that do not align with gene expression-based classifications.


