Fiora: Local neighborhood-based prediction of compound mass spectra from single fragmentation events
Fiora: Local neighborhood-based prediction of compound mass spectra from single fragmentation events
Nowatzky, Y.; Russo, F.; Lisec, J.; Kister, A.; Reinert, K.; Muth, T.; Benner, P.
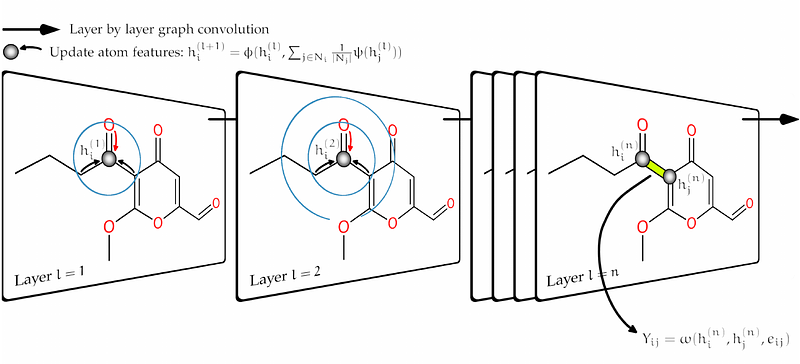
AbstractNon-targeted metabolomics holds great promise for advancing precision medicine and facilitating the discovery of novel biomarkers. However, the identification of compounds from tandem mass spectra remains a non-trivial task due to the incomplete nature of spectral reference libraries. Augmenting these libraries with simulated mass spectra can provide the necessary reference to resolve unmatched mass spectra, but remains a difficult undertaking to this day. In this study, we introduce Fiora, an innovative open-source algorithm using graph neural networks to simulate tandem mass spectra in silico. Our objective is to improve fragment intensity prediction with an intricate graph model architecture that facilitates edge prediction, thereby modeling fragment ions as the result of singular bond breaks and their local molecular neighborhood. We evaluate the performance on test data from NIST (2017) and the curated MS-Dial spectral library, as well as compounds from the 2016 and 2022 CASMI challenges. Fiora not only surpasses state-of-the-art fragmentation algorithms, ICEBERG and CFM-ID, in terms of prediction quality, but also predicts additional features, such as retention time and collision cross section. In addition, Fiora emonstrates significant speed improvements through the use of GPUs. This enables rapid (re)scoring of putative compound identifications in non-targeted experiments and facilitates large-scale expansion of spectral reference libraries with accurate spectral predictions.
