Deep Co-learning on transcription factors and their binding sites attains impeccable universality in plants
Deep Co-learning on transcription factors and their binding sites attains impeccable universality in plants
Gupta, S.; Kesarwani, V.; Bhati, U.; Jyoti, ; Shankar, R.
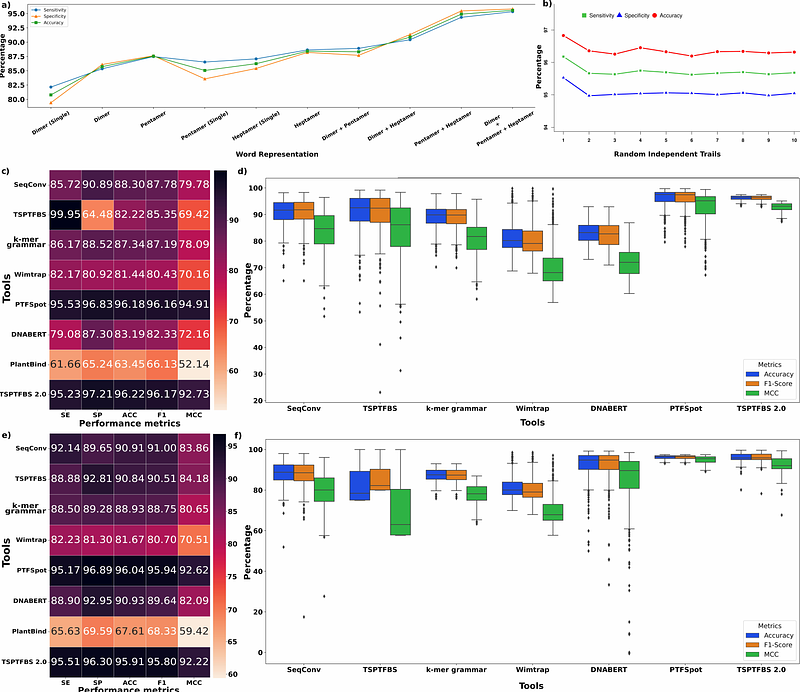
AbstractUnlike animals, variability in transcription factors (TF) and their binding sites (TFBS) across the plants is a major problem which most of the existing TFBS finding software fail to tackle, rendering them hardly of any use. This limitation has resulted into underdevelopment of plant regulatory research and rampant misuse of Arabidopsis models, generating misleading results. Here we report a ground-breaking transformers based deep-learning approach, PTFSpot, which learns from TF structures and their binding sites to bring a universal TF-DNA interaction model. During a series of extensive bench-marking studies, it not only outperformed the existing software by >20% lead, but also delivered consistently >90% accuracy even for those species and TF families which were never encountered during model building process. PTFSpot makes it possible now to accurately annotate TFBS across novel plant genomes even in the total lack of any TF information.