Faster and more accurate assessment of differential transcript expression with Gibbs sampling and edgeR 4.0
Faster and more accurate assessment of differential transcript expression with Gibbs sampling and edgeR 4.0
Baldoni, P. L.; Chen, L.; Smyth, G. K.
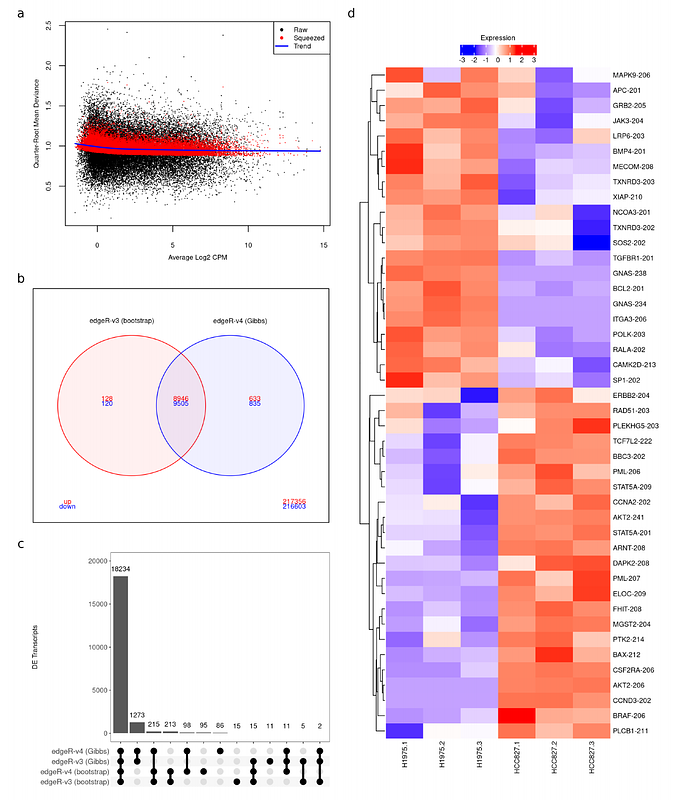
AbstractDifferential transcript expression analysis of RNA-seq data is becoming an increasingly popular tool to assess changes in expression of individual transcripts between biological conditions. Software designed for transcript-level differential expression analyses account for the uncertainty of transcript quantification, the read-to-transcript ambiguity (RTA), in statistical analyses via resampling methods. Bootstrap sampling is a popular resampling method that is implemented in the RNA-seq quantification tools kallisto and Salmon. However, bootstrapping is computationally intensive and provides replicate counts with low resolution when the number of sequence reads originating from a gene is low. For lowly expressed genes, bootstrap sampling results in noisy replicate counts for the associated transcripts, which in turn leads to non reproducible and unrealistically high RTA overdispersion for those transcripts. Gibbs sampling is a more efficient and high resolution algorithm implemented in Salmon. Here we leverage the latest developments of edgeR 4.0 to present an improved differential transcript expression analysis pipeline with Salmon\'s Gibbs sampling algorithm. The new bias-corrected quasi-likelihood method with adjusted deviances for small counts from edgeR, combined with the efficient Gibbs sampling algorithm from Salmon, provides faster and more accurate DTE analyses of RNA-seq data. Comprehensive simulations and test data show that the presented analysis pipeline is more powerful and efficient than previous differential transcript expression pipelines while providing correct control of the false discovery rate.


