How to quantify immigration from community abundance data using the Neutral Community Model
How to quantify immigration from community abundance data using the Neutral Community Model
Rafay, R.; Jones, E. W.; Sivak, D.; Fowler, J.
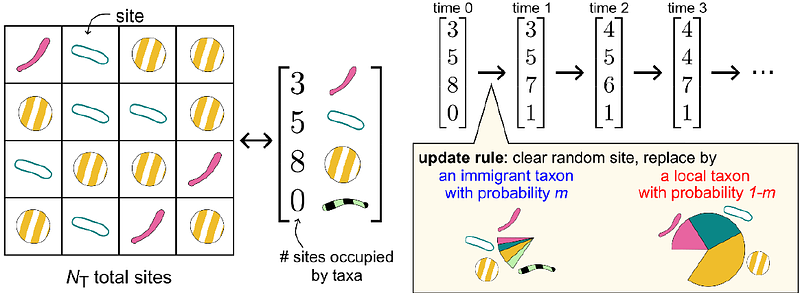
AbstractBiological communities are connected through dispersal, which regulates diversity across local and regional scales. However, dispersal is difficult to measure directly, limiting what is known about dispersal\'s impact on species composition in complex communities. One method to measure dispersal employs the Neutral Community Model (NCM) to quantify how a local community is influenced by the immigration of individuals from a larger source community. Conveniently, the immigration rate NTm of the NCM can be fit from biological sequence abundance datasets, which are plentiful. Yet it is neither known if these estimated values reflect the ground truth, nor what sampling effort is required to yield accurate estimates. In this study we introduce two inference methods, a variance-based and a Dirichlet-Multinomial Log-Likelihood (DM-LL) method, to complement the established occupancy-based inference method. In simulations of communities that resemble activated sludge microbiomes, all inference methods were capable of estimating NTm within 10% of ground-truth, with the variance-based and DM-LL methods requiring less sampling effort. Accurate inferences require read depths greater than NTm in each sample. The three methods agree in their inferred NTm in simulations of communities experiencing weak non-neutral effects (e.g., selection), and in applications to an empirical dataset from wastewater activated sludge. Based on these findings, we propose practical sampling and methodological guidelines for quantifying immigration between highly diverse, complex communities using the NCM.