Tracing SARS-CoV-2 Evolution in Algeria: Insights from 2020-2023
Tracing SARS-CoV-2 Evolution in Algeria: Insights from 2020-2023
Ezahedi, F. E.; Derrar, F.; Abraham, A.; Zeghbib, S.
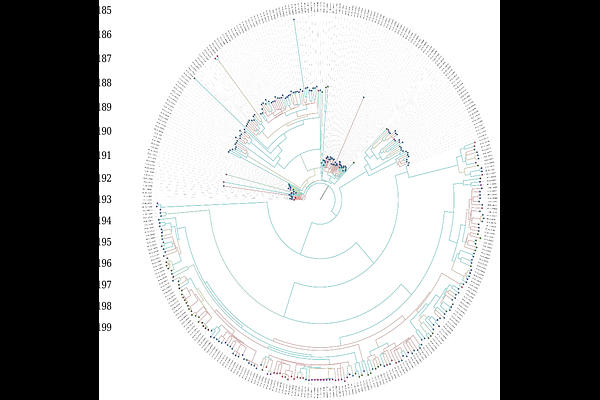
AbstractSARS-CoV-2 continues to circulate globally, leading to the emergence of new viral sublineages. This study examined 449 full-length genomic sequences from Algeria (March 2020 to May 2023) to explore SARS-CoV-2 evolution in this region. Our analysis revealed multiple introductions of viral strains, which were marked by significant genomic changes over time. Algeria-specific mutations were identified, providing insights into regional adaptations and evolutionary dynamics. Furthermore, a statistically significant recombination event was detected using the RDP 5 software. EPI_ISL_15920753 was recombinant, whereas EPI_ISL_15790700 and EPI_ISL_12156732 constituted the major and minor parents, respectively. Notably, haplotype analysis revealed 179 distinct haplotypes, including a highly connected node (H91) associated with a superspreading event later in the pandemic (July-September 2022), suggesting rapid transmission likely driven by mobile individuals and large gatherings. These findings emphasize the ongoing evolution of SARS-CoV-2 in local settings and highlight the importance of genomic surveillance for tracking emerging mutations and their potential impact.