SF3B1K700E Drives Oncogenic Alternative Splicing of Cell Cycle Checkpoint Genes in Cancer
SF3B1K700E Drives Oncogenic Alternative Splicing of Cell Cycle Checkpoint Genes in Cancer
Baker, M.; Sharma, A.; Engal, E.; Petasny, M.; Jaffe-Herman, S.; Geminder, O.; Su, M.; Bentata, M.; Kay, G.; Salton, M.
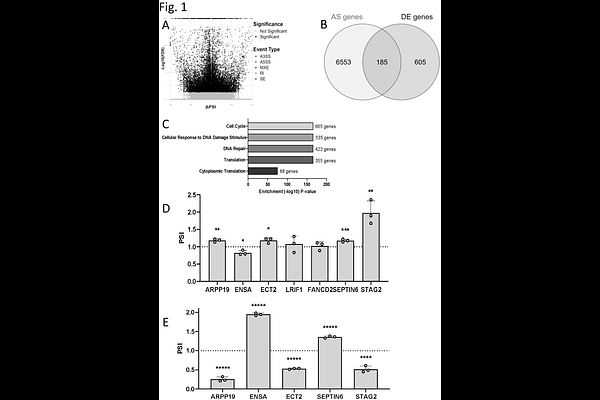
AbstractPre-mRNA splicing plays a crucial role in maintaining cellular homeostasis, with strict regulation required for processes such as cell cycle progression. SF3B1, a core component of the spliceosome, has emerged as a key player in alternative splicing regulation and is frequently mutated in cancer. Among these mutations, SF3B1K700E disrupts normal splicing patterns and deregulates cell cycle control. In this study, we investigated the impact of SF3B1 phosphorylation on cell cycle-dependent splicing, with a particular focus on the checkpoint regulators ARPP19 and ENSA, two functionally related proteins that modulate PP2A-B55 activity during mitotic progression. Using RNA-seq analysis, we identified a subset of SF3B1-regulated splicing events that are enriched in cell cycle genes. We demonstrated that SF3B1 phosphorylation during G2/M promotes the ARPP19-long isoform, a variant associated with poor survival in AML patients and accelerated mitotic exit. Notably, we have shown that inhibition of SF3B1 reverses the splicing changes induced by SF3B1K700E, reducing the levels of ARPP19-long, which suggests a potential therapeutic strategy for cancers with spliceosome mutations. Our findings highlight a direct link between SF3B1-dependent splicing, cell cycle progression, and tumorigenesis, providing new insights into the molecular mechanisms underlying cancer-associated splicing dysregulation.