Nanopore Direct RNA Sequencing Reveals Virus-Induced Changes in the Transcriptional Landscape in Human Bronchial Epithelial Cells
Nanopore Direct RNA Sequencing Reveals Virus-Induced Changes in the Transcriptional Landscape in Human Bronchial Epithelial Cells
Schroeder, S.; Wang, D.; Booth, J. L.; Wu, W.; Kiger, N.; Lettow, M.; Bates, A.; Pan, C.; Metcalf, J. P.
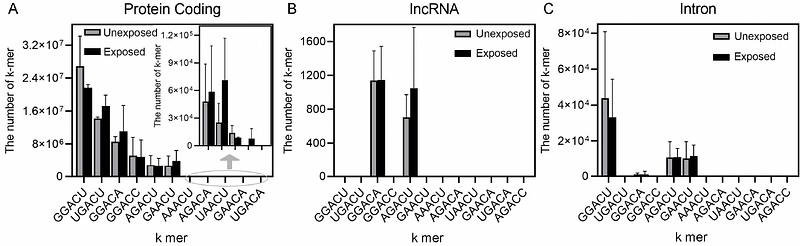
AbstractDirect RNA nanopore sequencing reveals changes in gene expression, polyadenylation, splicing, m6A methylation, and pseudouridylation in response to influenza virus exposure in primary human bronchial epithelial cells. This study focuses on the epitranscriptomic profile of genes in the host immune response. In addition to polyadenylated noncoding RNA, we purified and sequenced nonpolyadenylated noncoding RNA and observed changes in expression, N6-methyl-adenosine (m6A), and pseudouridylation ({Psi}) in these novel RNA. Two recently discovered lincRNA with roles in immune response, Chaserr and LEADR, became highly methylated in response to influenza exposure. Several H/ACA type snoRNAs that guide pseudouridylation are decreased in expression in response to influenza, and there is a corresponding decrease in the pseudouridylation of two novel lncRNA. Thus, novel epitranscriptomic changes revealed by direct RNA sequencing with nanopore technology provides unique insights into the host epitranscriptomic changes in epithelial gene networks that respond to influenza virus infection.


