Automated Discovery of Patterns in T-Cell Receptor Physicochemical Signatures
Automated Discovery of Patterns in T-Cell Receptor Physicochemical Signatures
Shams, Z.; Bishop, E.; Mckee-Reid, L.; Rumbelow, J.
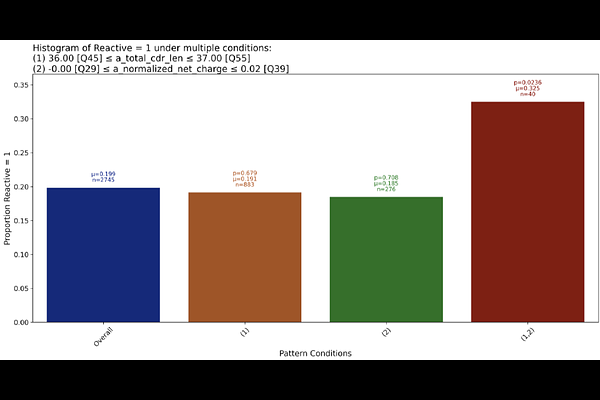
AbstractAccurately distinguishing antigen-reactive from non-reactive T-cell receptors (TCRs) is critical for advancing TCR-based immunotherapies and vaccines. Predicting antigen reactivity from physicochemical properties of the TCR sequence alone could enable rapid, low-cost identification of TCRs of interest, accelerating therapeutic discovery. In this paper, we use the Discovery Engine, a novel system for automated knowledge discovery from data, to classify published tumour antigen-reactive and non-reactive TCRs collected from cancer patients. Beyond classification, the Discovery Engine extracts interpretable combinatorial patterns (e.g., combinations of CDR3 length, net charge, and hydrophobicity) that predict whether a TCR is antigen-reactive. These patterns point to biologically meaningful features linked to tumour antigen recognition and could inform rational TCR design and prioritization. Notably, over half of the predictive patterns involve features from both the alpha and beta chains, highlighting the importance of considering both in assessing antigen specificity.