Structural features within precursor microRNA-20a regulate Dicer-TRBP processing
Structural features within precursor microRNA-20a regulate Dicer-TRBP processing
Liu, Y.; Harkner, C. T.; Westwood, M. N.; Munsayac, A.; Keane, S. C.
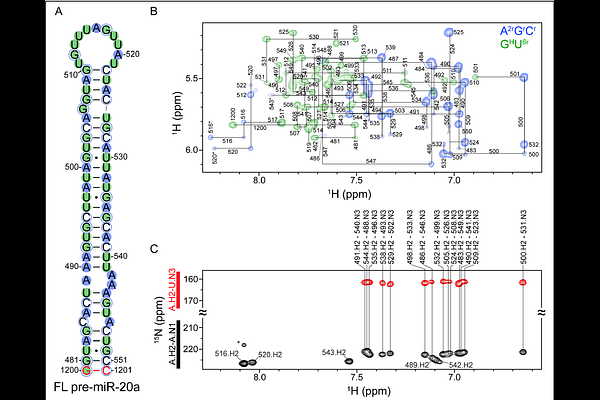
AbstractMicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression of target messenger (m) RNAs. To maintain proper miRNA expression levels, the enzymatic processing of primary and precursor miRNA elements must be strictly controlled. However, the molecular determinants underlying this strict regulation of miRNA biogenesis are not fully understood. Here, we determined the solution structure of pre-miR-20a, an oncogenic miRNA and component of the oncomiR-1 cluster, using nuclear magnetic spectroscopy (NMR) spectroscopy and small angle X-ray scattering (SAXS). Our structural studies informed on key secondary structure elements of pre-miR-20a which may control its enzymatic processing, namely a flexible apical loop and single-nucleotide bulge near the dicing site. We found that alternative conformations within pre-miR-20a\'s apical loop function to self-regulate its Dicer-TRBP processing, and that a single nucleotide bulge at the -5 position from the 5\'-cleavage site is critical for efficient processing. We additionally found that a disease-related single-nucleotide polymorphism in pre-miR-20a, predicted to disrupt the structure near the dicing site, resulted in reduced processing. These results further our structural understanding of the oncomiR-1 cluster and show how transient RNA conformers can function to self-regulate maturation.