Multiplexed tumor profiling with generative AI accelerates histopathology workflows and improves clinical predictions
Multiplexed tumor profiling with generative AI accelerates histopathology workflows and improves clinical predictions
Pati, P.; Karkampouna, S.; Bonollo, F.; Comperat, E.; Radic, M.; Spahn, M.; Martinelli, A.; Wartenberg, M.; Kruithof-de Julio, M.; Rapsomaniki, M. A.
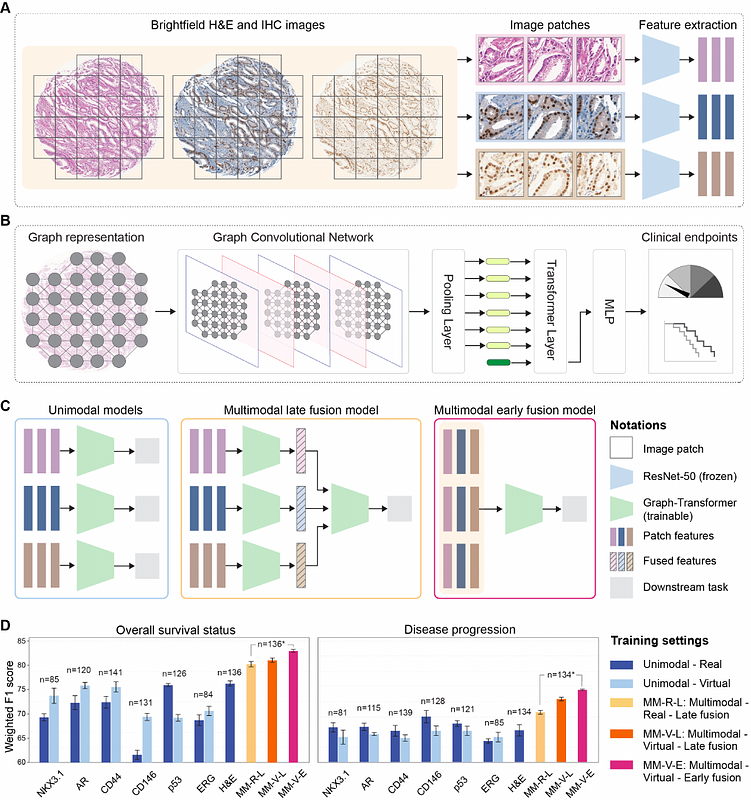
AbstractUnderstanding the spatial heterogeneity of tumors and its links to disease is a cornerstone of cancer biology. Emerging spatial technologies offer unprecedented capabilities towards this goal, but several limitations hinder their clinical adoption. To date, histopathology workflows still heavily depend on hematoxylin & eosin (H&E) and serial immunohistochemistry (IHC) staining, a cumbersome and tissue-exhaustive process that yields unaligned tissue images. We propose the VirtualMultiplexer, a generative AI toolkit that translates real H&E images to matching IHC images for several markers based on contrastive learning. The VirtualMultiplexer learns from unpaired H&E and IHC images and introduces a novel multi-scale loss to ensure consistent and biologically reliable stainings. The virtually multiplexed images enabled training a Graph Transformer that simultaneously learns from the joint spatial distribution of several markers to predict clinically relevant endpoints. Our results indicate that the VirtualMultiplexer achieves rapid, robust and precise generation of virtually multiplexed imaging datasets of high staining quality that are indistinguishable from the real ones. We successfully employed transfer learning to generate realistic virtual stainings across tissue scales, patient cohorts, and cancer types with no need for model fine-tuning. Crucially, the generated images are not only realistic but also clinically relevant, as they greatly improved the prediction of different clinical endpoints across patient cohorts and cancer types, speeding up histopathology workflows and accelerating spatial biology.