AptaBERT: Predicting aptamer binding interactions
AptaBERT: Predicting aptamer binding interactions
Morsch, F.; Umasankar, I. L.; Sanz Moreta, L.; Latawa, P.; Lange, D. B.; Wengel, J.; Konjen, H.; Code, C.
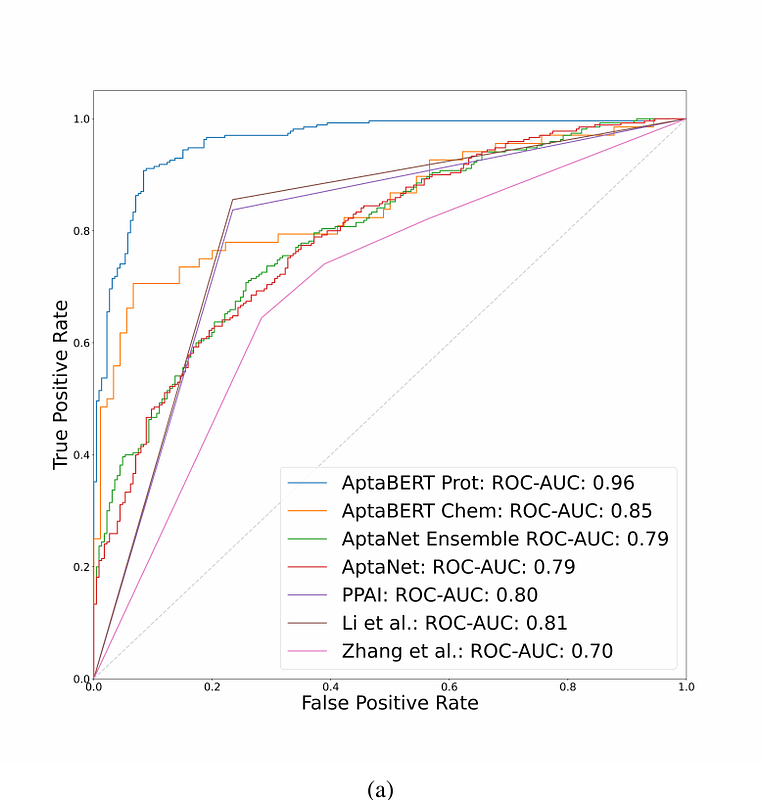
AbstractAptamers, short single-stranded DNA or RNA, are promising as future diagnostic and therapeutic agents. Traditional selection methods, such as the Systemic Evolution of Ligands by Exponential Enrichment (SELEX), are not without limitations being both resource-intensive and prone to biases in library construction and the selection phase. Leveraging Dianox\'s extensive aptamer database, we introduce a novel computational approach, AptaBERT, built upon the BERT architecture. This method utilizes self supervised pre-training on vast amounts of data, followed by supervised fine-tuning to enhance the prediction of aptamer interactions with proteins and small molecules. AptaBERT is fine-tuned for binary classification tasks, distinguishing between positive and negative interactions with proteins and small molecules. AptaBERT achieves an ROC-AUC of 96% for protein interactions, surpassing existing models by at least 15%. For small molecule interactions, AptaBERT attains a ROC-AUC of 85%. Our findings demonstrate AptaBERT\'s superior predictive capability and its potential to identify novel aptamers binding to targets.