Chemical representation standardization needed to generalize metabolic pathway involvement prediction across the Kyoto Encyclopedia of Genes and Genomes, Reactome, and MetaCyc knowledgebases
Chemical representation standardization needed to generalize metabolic pathway involvement prediction across the Kyoto Encyclopedia of Genes and Genomes, Reactome, and MetaCyc knowledgebases
Huckvale, E. D.; Moseley, H. N. B.
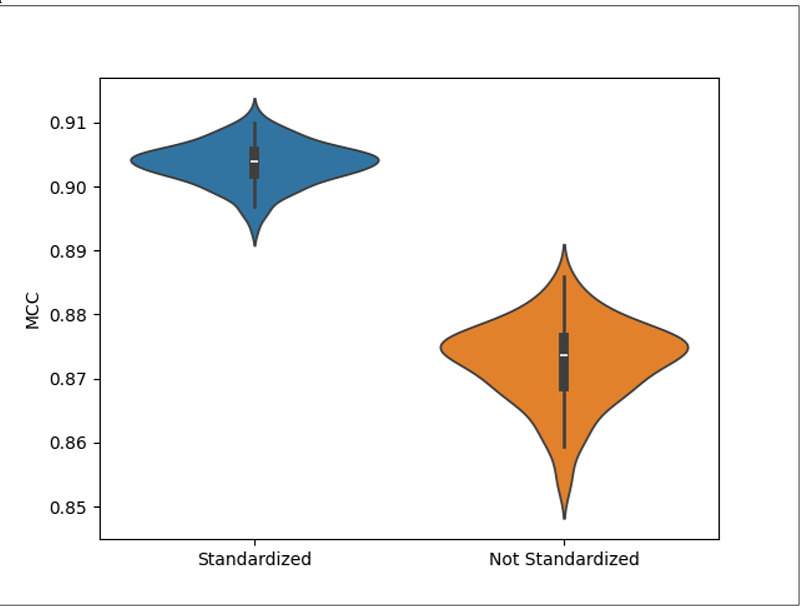
AbstractMotivation Due to the utility of knowing the pathway involvement of compounds detected in biological experiments, knowledgebases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome, and MetaCyc have aggregated pathway annotations of compounds. However, these annotations are largely incomplete and are costly to obtain experimentally and curate from published scientific literature. Results We constructed a new dataset using compounds and their pathway annotations from KEGG, Reactome, and MetaCyc. Using this dataset, we trained and tested an extreme classification model that classifies 8,195 unique pathways based on compound chemical representations with a mean Matthews correlation coefficient (MCC) of 0.9036 +/- 0.0033. During model evaluation, we discovered an inconsistency in chemical representations across knowledgebases, which was alleviated by standardizing the chemical representations using InChI (IUPAC International Chemical Identifier) canonicalization. Next, we compared the MCC between compounds and their cross-knowledgebase references. The non-standardized chemical representations had a huge 0.2687 drop in MCC while the standardized chemical representations only had a 0.0384 drop in MCC. Thus, standardizing chemical representation is an essential step when predicting on novel chemical representations.