Catalytic pocket-informed augmentation of enzyme kinetic parameters prediction via hierarchical graph learning
Catalytic pocket-informed augmentation of enzyme kinetic parameters prediction via hierarchical graph learning
Luo, D.; Qu, X.; Wang, B.
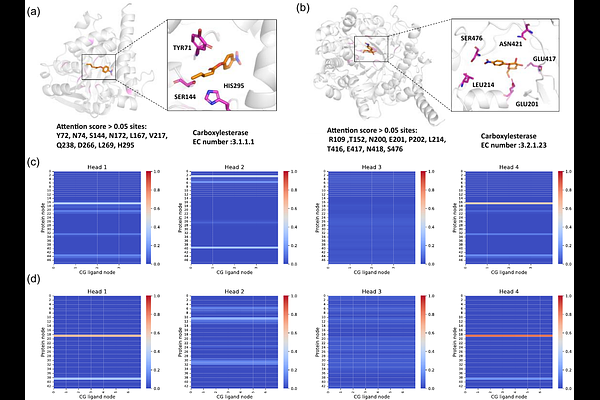
AbstractPredicting enzyme kinetic parameters is crucial for enzyme engineering and mining, but development of accurate tools for this task is still challenging due to the complexity of influencing factors. (LLMs)-based deep learning methods show satisfactory performance in Kcat prediction by encoding substrate and enzyme sequence information, but this further enhancement of the performance can be largely limited by the ignorance of the substrate-binding information. Here, we introduce GraphKcat, a deep learning framework that integrates enzyme-substrate 3D binding conformations for precise kinetic parameter prediction. The method employs Chai-1 to generate enzyme-substrate complex conformations, followed by a developed multi-scale hierarchical graph neural network to systematically characterize active site features from all-atom (AA) level to coarse-grained (CG) level, which are subsequently fused with LLMs embedded substrate, sequence, and environmental related information for prediction. To enable efficient multimodal feature integration, we proposed a multi modal cross-attention fusion (MMCAF) module for feature alignment and update. Experimental evaluations demonstrate that GraphKcat can effectively identifying catalysis-critical residues, learning sequence similarity-independent features through its multi-scale graph architecture, and maintaining robust performance under low sequence similarity conditions. Critically, GraphKcat captures conserved structural and physicochemical patterns related to enzymatic activity, enabling accurate identification of high-activity enzyme variants and functional mutants even for low-homology targets. These capabilities highlight its potential as a transformative tool for enzymatic activity prediction, rational enzyme engineering, and enzyme mining in industrial applications.