Benchmarking single cell transcriptome matching methods for incremental growth of reference atlases
Benchmarking single cell transcriptome matching methods for incremental growth of reference atlases
Hu, J.; Peng, B.; Pankajam, A. V.; Xu, B.; Deshpande, V. A.; Bueckle, A. D.; Herr, B. W.; Borner, K.; Dupont, C. L.; Scheuermann, R. H.; Zhang, Y.
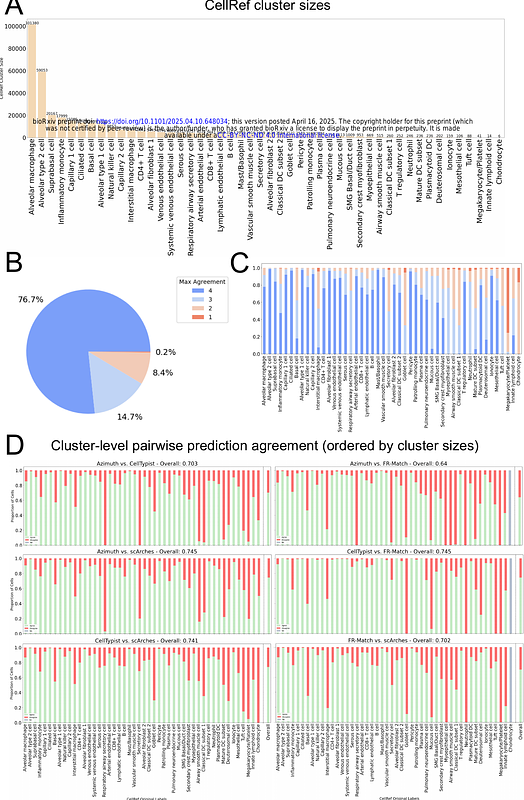
AbstractBackground: The advancement of single cell technologies has driven significant progress in constructing a multiscale, pan-organ Human Reference Atlas (HRA), though challenges remain in harmonizing cell types and unifying nomenclature. Multiple machine learning and artificial intelligence methods, including pre-trained and fine-tuned models on large-scale atlas data, are publicly available for the single cell community users to computationally annotate and match their cell clusters to the reference atlas. Results: This study benchmarks four computational tools for cell type annotation and matching -- Azimuth, CellTypist, scArches, and FR-Match -- using two lung atlas datasets, the Human Lung Cell Atlas (HLCA) and the LungMAP single-cell reference (CellRef). Despite achieving high overall performance while comparing algorithmic cell type annotations to expert annotated data, variations in accuracy were observed, especially in annotating rare cell types, underlining the need for improved consistency across cell type prediction methods. The benchmarked methods were used to cross-compare and incrementally integrate 61 cell types from HLCA and 48 cell types from CellRef, resulting in a meta-atlas of 41 matched cell types, 20 HLCA-specific cell types, and 7 CellRef-specific cell types. Conclusion: This study reveals complementing strengths of the benchmarked methods and presents a framework for incremental growth of the cell type inventory in the reference atlases, leading to 68 unique cell types in the meta-atlas across CellRef and HLCA. The benchmarking analysis contributes to improving the coverage and quality of HRA construction by assessing the reliability and performance of cell type annotation approaches for single cell transcriptomics datasets.