Explainable Multimodal Machine Learning Using Combined Environmental DNA and Biogeographic Features for Ecosystem Biomonitoring
Explainable Multimodal Machine Learning Using Combined Environmental DNA and Biogeographic Features for Ecosystem Biomonitoring
Koh, J. C. O.
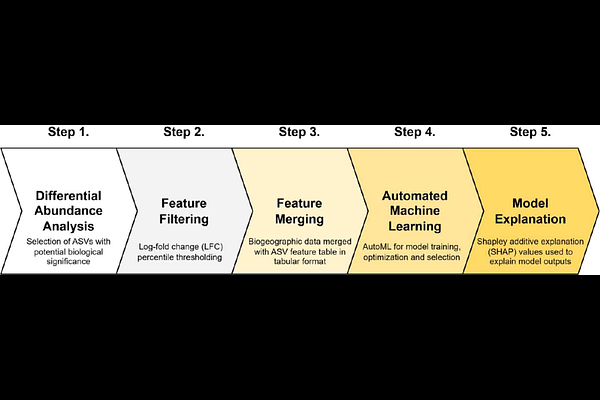
AbstractMachine learning (ML) has been proposed as a taxonomy-independent approach using environmental DNA (eDNA) for ecosystem biomonitoring. Representations of eDNA amplicons either as clustered sequences termed operational taxonomic units (OTUs) or unique sequences termed amplicon sequence variants (ASVs) are used as inputs in current ML practices, with varied successes reported. Biogeographic data encompassing the physical, climate and ecological observations collected at biomonitoring sites provide a repository of potential informative features that can augment ML performance in combination with eDNA data. I introduce a multimodal ML workflow using combined eDNA and biogeographic features for ecosystem biomonitoring. Differentially abundant ASVs were merged with biogeographic data and used as input in an automated ML approach. Using Switzerland\'s freshwater macroinvertebrate eDNA dataset collected across 163 biomonitoring sites and impact prediction as an example, I show that the multimodal ML approach (83.3% accuracy) significantly outperformed ML using only ASVs (66.7% accuracy) or OTUs (64.6% accuracy). Shapley additive explanation of the best ML model revealed key biogeographic features and species/taxa impacting upon predictions. The proposed workflow can be readily adopted in existing bioinformatics/ML pipelines and will further advance the use of eDNA for biomonitoring.