Scalable and cost-efficient custom gene library assembly from oligopools
Scalable and cost-efficient custom gene library assembly from oligopools
Freschlin, C.; Yang, K.; Romero, P. A.
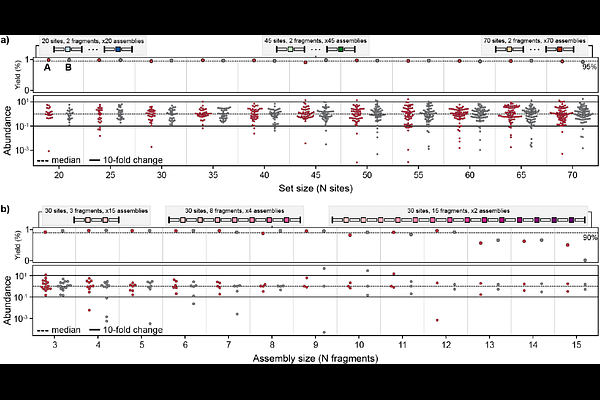
AbstractAdvances in metagenomics, deep learning, and generative protein design have enabled broad in silico exploration of sequence space, but experimental characterization is still constrained by the cost and scalability of DNA synthesis. Here, we present OMEGA (Oligo-based Multiplexed Efficient Gene Assembly), a low-cost, accessible method for assembling hundreds to thousands of full-length genes in parallel using standard laboratory techniques. OMEGA computationally fragments target genes into short, high-fidelity Golden Gate-compatible oligonucleotides that can be ordered as a pooled library and assembled across multiplexed subpools. We systematically optimized the number of fragments per gene and orthogonal ligation sites per reaction and determine that OMEGA can assemble up to 2.6 kb constructs using as many as 70 Golden Gate sites. To validate the approach, we assembled and functionally screened a library of 810 natural and synthetic GFP variants, recovering 94-97% of target sequences with high uniformity. OMEGA enables precision library construction at scale, with per-gene costs as low as $1.50, and offers a broadly applicable solution for bridging computational protein design with high-throughput experimental validation.