Dichotomous transcriptome divergence at the cell-type level between the Drosophila male and female germlines
Dichotomous transcriptome divergence at the cell-type level between the Drosophila male and female germlines
Hariyani, I. E.; Das, S.; Le, E. M.; Gamero-Castano, C.; Soroudi, T.; Choi, J.; Swarup, V.; Ranz, J. M.
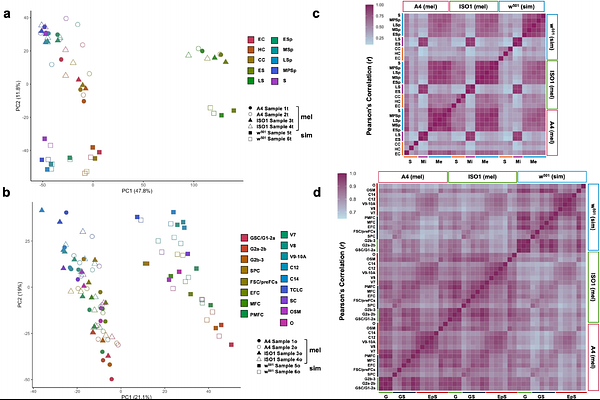
AbstractGene expression evolution in reproductive organs plays a central role in species divergence, yet cell-type-resolved patterns in invertebrates remain poorly understood. We used single-nucleus RNA-sequencing to profile testis and ovary transcriptomes from the sibling species Drosophila melanogaster and D. simulans. Despite conserved cellular composition, we observed highly cell-type-specific expression and coexpression network divergence between species. In the testis, mitotic cells were conserved across species, mirroring patterns in mammals, while divergence peaked in late spermatocytes. In the ovary, divergence was less pronounced, peaking in early germline and late-stage follicle cells, and enriched on the X chromosome, consistent with a faster-X effect driven by positive selection. Genes expressed in these cell types exhibited narrower expression breadth, younger phylogenetic age, and elevated rates of protein evolution in both tissues. Our findings reveal contrasting evolutionary regimes in male and female germlines, shaped by adaptive and non-adaptive mechanisms, contingent on cell type and chromosomal context.