DeepEpitope: Leveraging Transformation-Based protein Embeddings for Accurate linear Cancer B-cell Epitope Identification
DeepEpitope: Leveraging Transformation-Based protein Embeddings for Accurate linear Cancer B-cell Epitope Identification
T, D.; Rao, P. V.; Vasudevan, K.
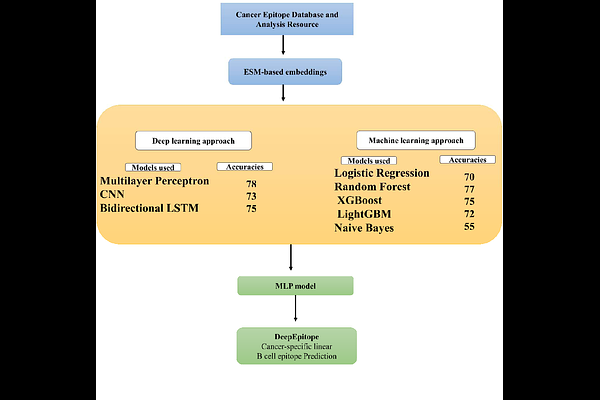
AbstractConventional cancer treatments tend to have serious side effects, leading to the quest for safer and more specific treatment modalities. Immunotherapy with vaccines has appeared as a promising option, with B-cell epitopes being crucial for the generation of humoral immunity. But identification of the correct B-cell epitopes of cancer is a severe challenge since current tools are not pre-trained with cancer-generated datasets. To bridge this gap, we introduce DeepEpitope, a command-line tool based on deep learning designed exclusively for the prediction of linear B-cell epitopes from cancer antigens. We compiled a high-quality dataset from the Cancer Epitope Database and used Evolutionary Scale Modeling (ESM) embeddings to represent epitope and non-epitope sequences as vectors of 1280 dimensions. These embeddings were employed to train five machine learning models (Logistic Regression, Random Forest, XGBoost, LightGBM, and Naive Bayes) and three deep learning models (Multilayer Perceptron [MLP], Convolutional Neural Network, and Bidirectional LSTM). Of these, the MLP model performed best with an AUC of 0.85 and a benchmark AUC of 94%. In comparison with other tools like BepiPred (60%) and LBtope (54%), DeepEpitope demonstrated much higher predictive accuracy. It is a Linux-based command-line tool that can be accessed for free at: https://github.com/karthick1087/DeepEpitope.