Evidence-driven biases in alternative splicing inferred from NCBI Eukaryotic Genome Annotation Pipeline metadata
Evidence-driven biases in alternative splicing inferred from NCBI Eukaryotic Genome Annotation Pipeline metadata
de la Fuente, R.; Diaz-Villanueva, W.; Arnau, V.; Moya, A.
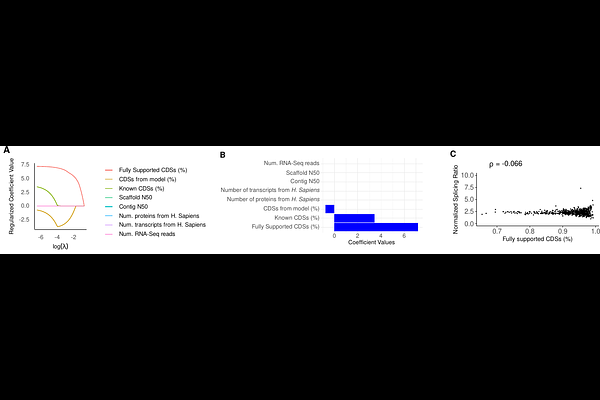
AbstractThe NCBI Eukaryotic Genome Annotation Pipeline (EGAP) predicts coding sequences by integrating transcriptomic and proteomic data with computational approaches, providing structural information that can be used to infer alternative transcript isoforms. However, accurate estimation of alternative splicing events depends on high-quality genome annotations, particularly in genome-wide analyses. The extent to which annotation pipelines influence these inferred splicing patterns has remained largely unexplored. In this study, we quantify potential biases associated with the EGAP annotation pipeline, and find that specific annotation features strongly influence these estimates, particularly the percentage of coding sequences that are supported by experimental evidence. Further, we implemented a polynomial regression model to normalize splicing levels, generating an adjusted metric that minimizes evidence-driven biases. This framework may serve as a basis for future investigations into splicing complexity and comparative genomics.