From sequence to molecules: Feature sequence-based genome mining uncovers the hidden diversity of bacterial siderophore pathways
From sequence to molecules: Feature sequence-based genome mining uncovers the hidden diversity of bacterial siderophore pathways
Gu, S.; Shao, Y.; Rehm, K.; Bigler, L.; Zhang, D.; He, R.; Shao, J.; Jousset, A.; Friman, V.-P.; Wei, Z.; Kummerli, R.; Li, Z.
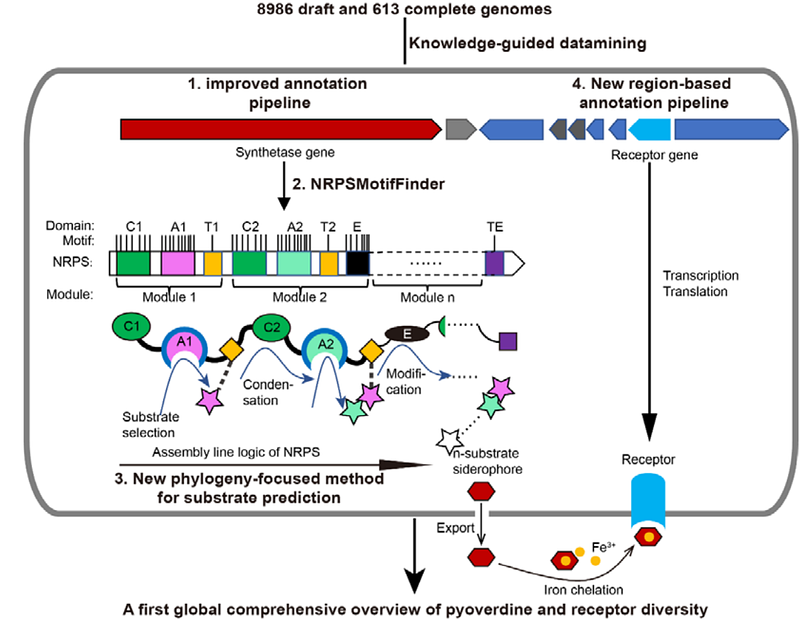
AbstractMicrobial secondary metabolites have long been recognized as a rich source for pharmaceutical compound discovery and to have crucial ecological functions. However, the sequence-to-function mapping in microbial secondary metabolism pathways remains challenging because neither protein function nor substrate specificity can accurately be predicted from genome data. Here we focus on the iron-scavenging pyoverdines, siderophores of Pseudomonas bacteria, as model system to develop a knowledge-guided bioinformatic pipeline that extracts functional information of both the pyoverdine synthesis machinery and uptake receptors from 1928 draft genomes. For pyoverdine synthesis, our approach predicts the chemical structure of 188 different pyoverdines with nearly 100% accuracy. For pyoverdine uptake, our pipeline uncovers 94 different pyoverdine receptor groups. Our results demonstrate that combining feature sequence and phylogenetic approaches is a powerful way to reconstruct bacterial secondary metabolism pathways based on sequence data, unveiling an enormous yet overlooked diversity of siderophores and their receptors.