Defining the chromatin-associated protein landscapes on Trypanosoma brucei repetitive elements using synthetic TALE proteins
Defining the chromatin-associated protein landscapes on Trypanosoma brucei repetitive elements using synthetic TALE proteins
Carloni, R.; Devlin, T.; Tong, P.; Auchynnikava, T.; Spanos, C. R.; Rappsilber, J.; Matthews, K. R.; Allshire, R. C.
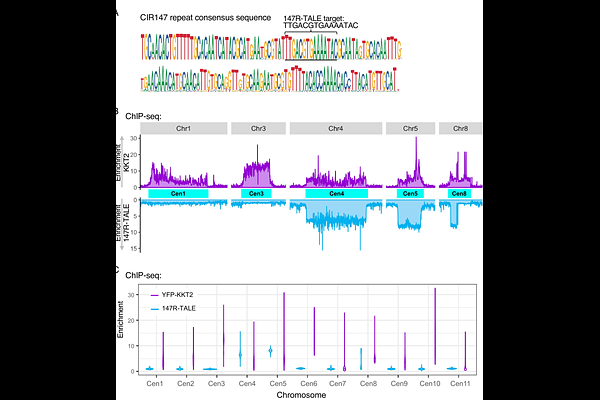
AbstractKinetoplastids, such as Trypansoma brucei, are eukaryotes that likely separated from the main lineage at an exceptionally early point in evolution. Consequently, many aspects of kinetoplastid biology differ significantly from other eukaryotic model systems, including yeasts, plants, worms, flies and mammals. As in many eukaryotes the T. brucei genome contains repetitive elements at various chromosomal locations including centromere- and telomere-associated repeats and interspersed retrotransposon elements. T. brucei also contains intermediate-sized and mini-chromosomes that harbor abundant 177 bp repeat arrays, and 70 bp repeat elements implicated in Variable Surface Glycoprotein (VSG) gene switching. In many eukaryotes repetitive elements are assembled in specialised chromatin such as heterochromatin, however, apart from centromere- and telomere-associated repeats, little is known about chromatin-associated proteins that decorate these and other repetitive elements in kinetoplastids. Here we utilize affinity selection of synthetic TALE DNA binding proteins designed to target specific repeat elements to identify enriched proteins by proteomics. Validating the approach, a telomere repeat binding TelR-TALE identifies many proteins previously implicated in telomere function. Furthermore, the 70R-TALE designed to bind 70 bp repeats indicates that proteins involved in DNA repair are enriched on these elements that reside adjacent to VSG genes. Interestingly, the 177 bp repeat binding 177R-TALE enriches for many kinetochore proteins suggesting that intermediate-sized and mini- chromosomes assemble kinetochores related in composition to those located on the main megabase chromosomes. This provides a first insight into the chromatin landscape of repetitive regions of the trypanosome genome with relevance for their mechanisms of chromosome integrity, immune evasion and cell replication.