Genomic tailoring of autogenous poultry vaccines to reduce Campylobacter from farm to fork
Genomic tailoring of autogenous poultry vaccines to reduce Campylobacter from farm to fork
Calland, J. K.; Pesonen, M. E.; Mehat, J.; Pascoe, B.; Haydon, D. J.; Lourenco, J.; Mourkas, E.; Hitchings, M. D.; LaRagione, R. M.; Hammond, P.; Wallis, T. S.; Corander, J.; Sheppard, S. K.
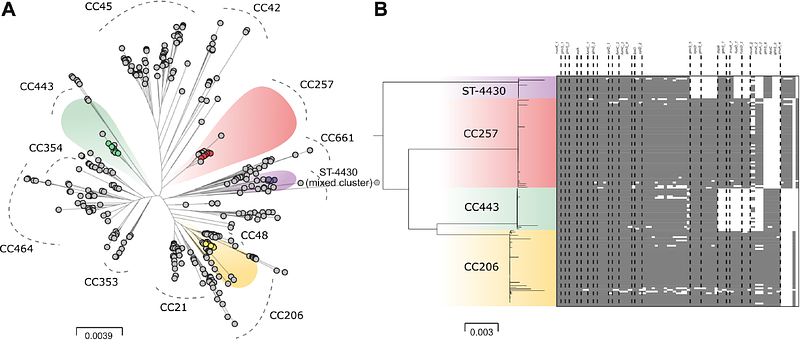
AbstractCampylobacter is a leading cause of food-borne gastroenteritis worldwide, linked to the consumption of contaminated poultry meat. Targeting this pathogen at source, vaccines for poultry can provide short term caecal reductions in Campylobacter numbers in the chicken intestine. However, this approach is unlikely to reduce Campylobacter in the food chain or human incidence. This is likely as vaccines typically target only a subset of the high strain diversity circulating among chicken flocks and rapid evolution diminishes vaccine efficacy over time. To address this, we used a genomic approach to develop a whole-cell autogenous vaccine targeting isolates harbouring genes linked to survival outside of the host. We hyper-immunised a whole major UK breeder farm to passively target offspring colonisation using maternally-derived antibody. Monitoring progeny, broiler flocks revealed a near-complete shift in the post-vaccination Campylobacter population with a ~50% reduction in isolates harbouring extra-intestinal survival genes and a significant reduction of Campylobacter cells surviving on the surface of meat. Based on these findings, we developed a logistic regression model that predicted that vaccine efficacy could be extended to target 46% of a population of clinically relevant strains. Immuno-manipulation of poultry microbiomes towards less harmful commensal isolates by competitive exclusion, has major potential for reducing pathogens in the food production chain.