The complexity of multiple CRISPR arrays in strains with (co-occurring) CRISPR systems
The complexity of multiple CRISPR arrays in strains with (co-occurring) CRISPR systems
Fehrenbach, A.; Mitrofanov, A.; Backofen, R.; Baumdicker, F.
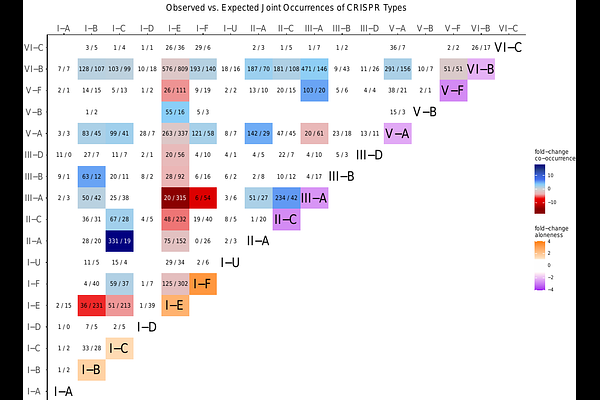
AbstractCRISPR and their associated Cas proteins provide adaptive immunity in prokaryotes, protecting against invading genetic elements. These systems are categorized into types and are highly diverse. Genomes often harbor multiple CRISPR arrays varying in length and distance from Cas loci. However, the ecological roles of multiple CRISPR arrays and their interactions with multiple Cas loci remain poorly understood. We present a comprehensive analysis of CRISPR systems that uncovers variation between diverse Cas types regarding the occurrence of multiple arrays, the distribution of their lengths and positions relative to Cas loci, and the diversity of their repeat sequences. Some types tend to occurr as the sole Cas present, but typically comprise two or more arrays, especially for types I-E and I-F. Multiple Cas types are also common, with some systems showing a preference for specific co-occurrence. Distinct array distributions and orientations around Cas loci indicate substantial differences in functionality and transcriptional behavior among Cas types. Our analysis suggests that arrays with identical repeats in the same genome acquire new spacers at comparable rates, irrespective of their proximity to the Cas locus. Furthermore, repeat similarities in our data set indicate that arrays of systems that often co-occur with other systems tend to have more diverse repeats than those mostly appearing alongside solitary systems within the genome. Our analysis suggests that co-occurring Cas type pairs might not only collaborate in spacer acquisition but also maintain independent and complementary functions and that CRISPR systems distribute their defensive spacer repertoire equally across multiple CRISPR arrays.