Differential expression of core metabolic functions in Candidatus Altiarchaeum inhabiting distinct subsurface ecosystems
Differential expression of core metabolic functions in Candidatus Altiarchaeum inhabiting distinct subsurface ecosystems
Esser, S. P.; Turzynski, V.; Plewka, J.; Moore, C. J.; Banas, I.; Soares, A.; Lee, J.; Woyke, T.; Probst, A. J.
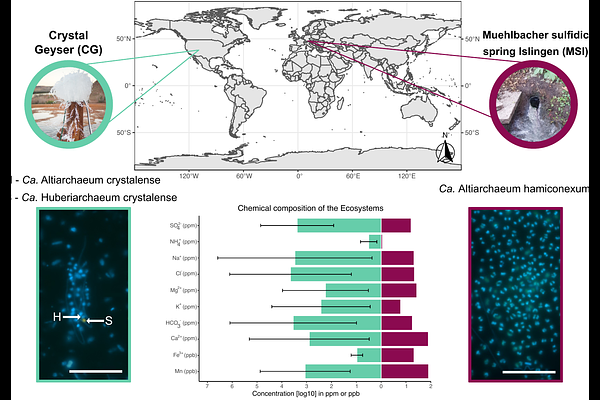
AbstractCandidatus Altiarchaea are widespread across aquatic subsurface ecosystems and possess a highly conserved core genome, yet adaptations of this core genome to different biotic and abiotic factors based on gene expression remain unknown. Here, we investigated the metatranscriptome of two Ca. Altiarchaeum populations that thrive in two substantially different subsurface ecosystems. In Crystal Geyser, a high-CO2 groundwater system in the USA, Ca. Altiarchaeum crystalense co-occurs with the symbiont Ca. Huberiarchaeum crystalense, while in the Muehlbacher sulfidic spring in Germany, an artesian spring high in sulfide concentration, Ca. A. hamiconexum is heavily infected with viruses. We here mapped metatranscriptome reads against their genomes to analyze the in situ expression profile of their core genomes. Out of 537 shared gene clusters, 331 were functionally annotated and 130 differed significantly in expression between the two sites. Main differences were related to genes involved in cell defense like CRISPR-Cas, virus defense, replication, and transcription as well as energy and carbon metabolism. Our results demonstrate that altiarchaeal populations in the subsurface are likely adapted to their environment while influenced by other biological entities that tamper with their core metabolism. We consequently posit that viruses and symbiotic interactions can be major energy sinks for organisms in the deep biosphere.