Spatial Transcriptomics Using Archived Formalin-Fixed Paraffin-Embedded Core Needle Biopsy Tissues Revealed Unique Transcriptomic Signatures in Kidney Transplant Rejections
Spatial Transcriptomics Using Archived Formalin-Fixed Paraffin-Embedded Core Needle Biopsy Tissues Revealed Unique Transcriptomic Signatures in Kidney Transplant Rejections
Wongworawat, Y. C.; Nepal, C.; Duhon, M.; Chen, W.; Nguyen, M.-T.; Godzik, A.; Qiu, X.; Li, V. W.; Yu, G.; Villicana, R.; Zuppan, C.; De Vera, M.; Haas, M.; Wang, C.
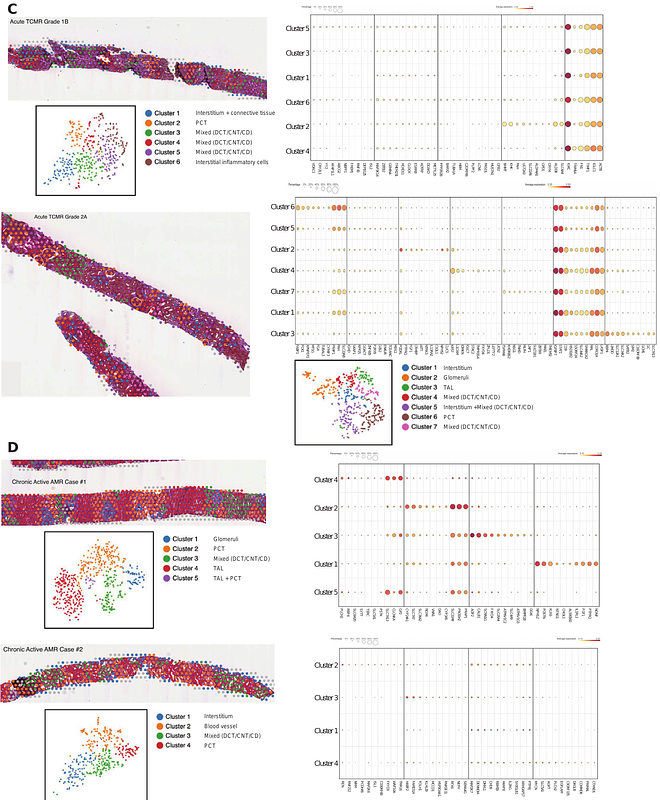
AbstractClinical core needle biopsies present challenges for transcriptomic analysis due to limited tissue volume. In this study, we determined the feasibility of using spatial transcriptomics to evaluate rejection on formalin-fixed paraffin-embedded (FFPE) core needle biopsies from human kidney allografts. We demonstrated that non-rejection, active antibody mediated rejection (AMR), acute cell mediated rejection (TCMR) and chronic active AMR have distinct transcriptomic signatures. Subclusters of monocytes/macrophages with high Fc gamma receptor IIIA (FCGR3A) expression were identified in C4d-positive active AMR and acute TCMR, and the spatial distribution of these cells corresponded to the characteristic histopathological features. Key markers related to monocyte/macrophage activation and innate alloantigen recognition were upregulated, along with metabolic pathways associated with trained immunity in AMR and TCMR. The discovery of unique transcriptomic signatures associated with AMR and TCMR facilitates the differentiation of acute kidney allograft rejections using spatial transcriptomic data derived from FFPE core needle biopsies.