Orthologs of an essential orphan gene vary in their capacities for function and subcellular localization in Drosophila melanogaster
Orthologs of an essential orphan gene vary in their capacities for function and subcellular localization in Drosophila melanogaster
Patel, P. H.; Eicholt, L. A.; Lange, A.; McDermott, K. L.; Bornberg-Bauer, E.; Findlay, G. D.
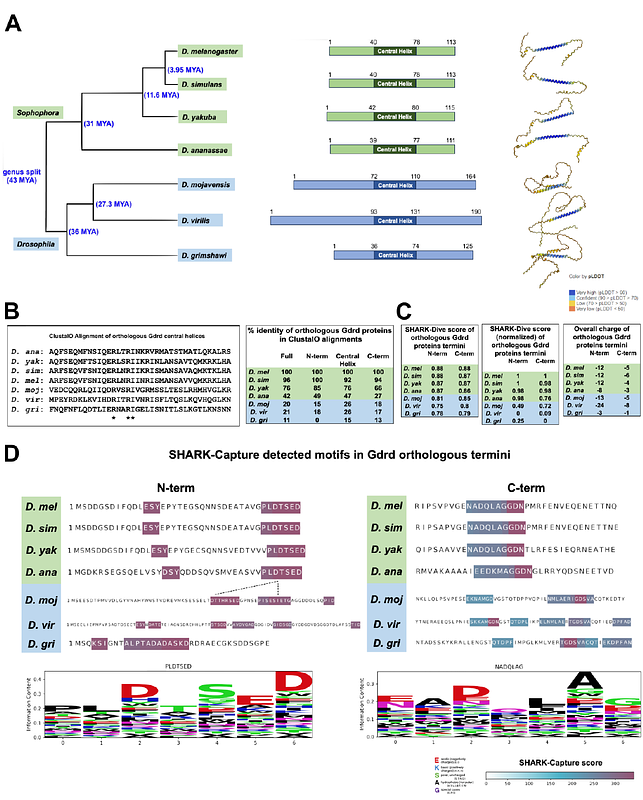
AbstractThe rapidly evolving nature of orphan genes raises questions about whether their activities remain conserved or evolve uniquely across species. We explore this issue using goddard (gdrd), an orphan gene essential for spermatogenesis in Drosophila melanogaster. Although the Gdrd protein has maintained structural conservation across the Drosophila genus, its overall length and primary sequences, especially at its disordered termini, show substantial divergence. Using gene swaps assays that included gdrd orthologs from multiple species across different evolutionary distances, we investigated how lineage-specific evolutionary changes have affected Goddard\'s ability to function in D. melanogaster. Our results indicate that most orthologs possess the ability to interact with axonemes and insect ring centrioles in D. melanogaster, suggesting these interactions could have been present in the orthologs\' common ancestor. Surprisingly, a highly divergent ortholog from D. mojavensis fully rescued fertility in goddard null D. melanogaster, indicating that the Gdrd protein at the base of the Drosophila genus was most likely fully integrated into an essential spermatogenesis pathway. However, several orthologs, including one from a more closely related species, failed to complement fertility in D. melanogaster. All of these complementation failures correlated with weaker axonemal binding. Furthermore, we noted that all tested orthologs exhibited divergent subcellular localization patterns. In parallel with these gene swap studies, we computationally analyzed each ortholog\'s structural stability and flexibility, physicochemical properties within its intrinsically disordered regions, and amino acid sequence divergence. Molecular dynamics simulations indicated structural instability in the most closely related, non-functional ortholog, while shared motifs identified in the disordered termini correlated with functionality, notably present in D. mojavensis and D. melanogaster but absent in non-rescuing orthologs. These findings suggest that although Goddard\'s central structure and key interactions likely existed at the base of the Drosophila genus, several orthologs have undergone consequential lineage-specific evolutionary changes.