Prime editor-based high-throughput screening reveals functional synonymous mutations in the human genome
Prime editor-based high-throughput screening reveals functional synonymous mutations in the human genome
Niu, X.; Tang, W.; Liu, Y.; Liu, Y.; Mo, B.; Yu, Y.; Wei, W.
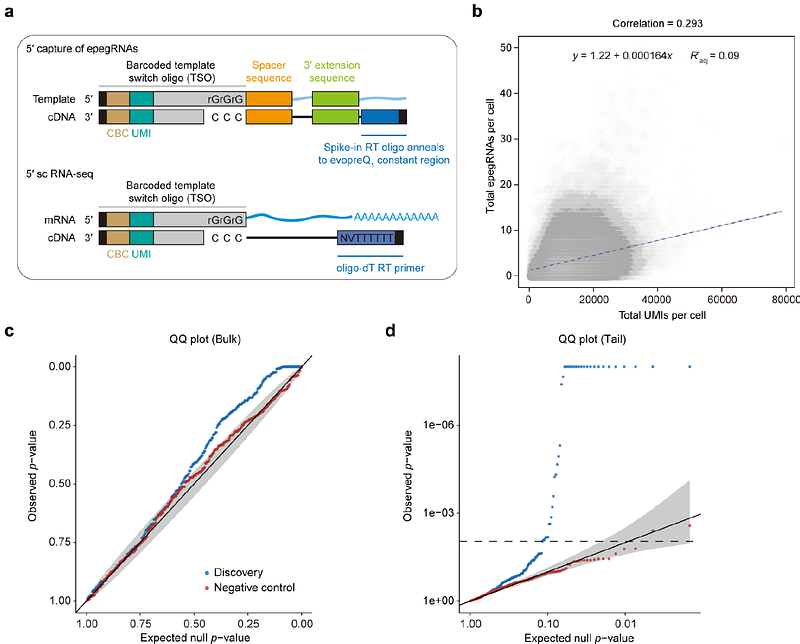
AbstractSynonymous mutations are generally considered neutral, while their roles in the human genome remain largely unexplored. Herein, we employed the PEmax system to create a library of 297,900 epegRNAs and performed extensive screening to identify synonymous mutations that impact cell fitness. While most synonymous mutations are neutral, we found that some can elicit phenotypic effects. By developing a specialized machine learning tool, we uncovered their impact on various biological processes such as mRNA splicing and transcription, supported by multifaceted experimental evidence. Notably, synonymous mutations can alter RNA folding and affect translation, as demonstrated by PLK1_S2. By integrating screening data with our model, we successfully predicted clinically deleterious synonymous mutations. This research deepens our understanding of synonymous mutations and provides insights for clinical disease studies.