How repeats rearrange chromosomes in deer mice
How repeats rearrange chromosomes in deer mice
Gozashti, L.; Harringmeyer, O. S.; Hoekstra, H. E.
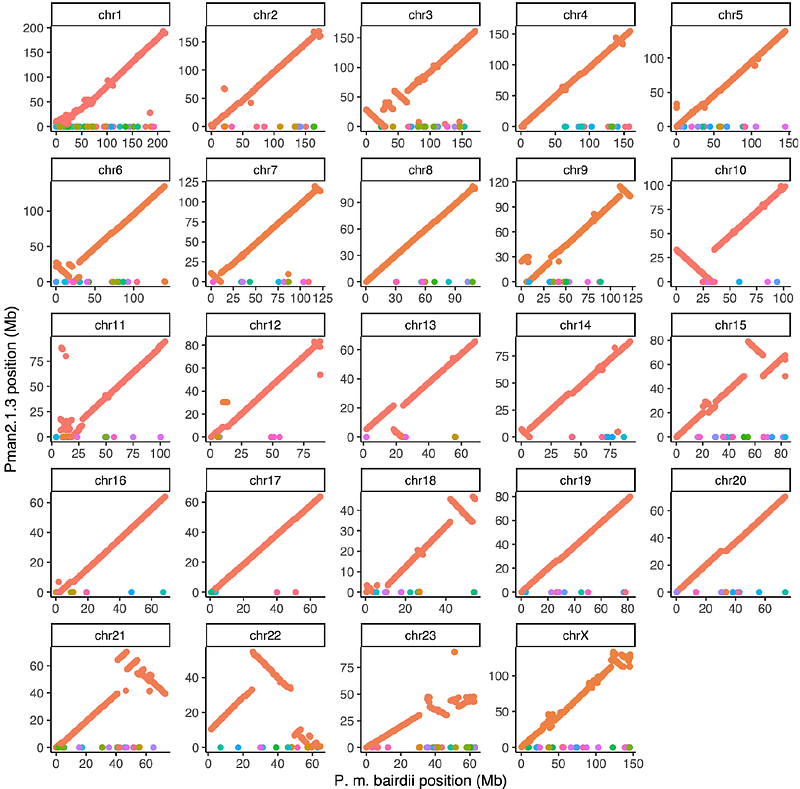
AbstractLarge genomic rearrangements, such as chromosomal inversions, can play a key role in evolution and often underlie karyotype variation, but the mechanisms by which these rearrangements arise remain poorly understood. To study the origins of inversions, we generated chromosome-level de novo genome assemblies for four subspecies of deer mice (Peromyscus maniculatus) with known inversion polymorphisms. We identified ~8,000 inversions, including 47 mega-base scale inversions, that together affect ~30% of the genome. Analysis of inversion breakpoints suggests that while most small (<1 Mb) inversions arise via ectopic recombination between retrotransposons, large (>1 Mb) inversions are primarily associated with segmental duplications (SDs). Large inversion breakpoints frequently occur near centromeres, which may be explained by an accumulation of transposable elements in pericentromeric regions driving SD formation. Additionally, multiple large inversions likely arose from ectopic recombination between near-identical centromeric satellite arrays located megabases apart, a previously uncharacterized mechanism of inversion formation. Together, our results illuminate how repeats give rise to massive shifts in chromosome architecture.