Persistently increased expression of PKMzeta and unbiased gene expression profiles identify hippocampal molecular traces of a long-term active place avoidance memory and 'shadow' proteins
Persistently increased expression of PKMzeta and unbiased gene expression profiles identify hippocampal molecular traces of a long-term active place avoidance memory and 'shadow' proteins
Han, J.; Grau-Perales, A.; Harris, R. M.; Kao, H.-y.; Pal, A.; Alarcon, J.-M.; Sacktor, T. C.; Martiniani, S.; Hofmann, H. A.; Fenton, A. A.
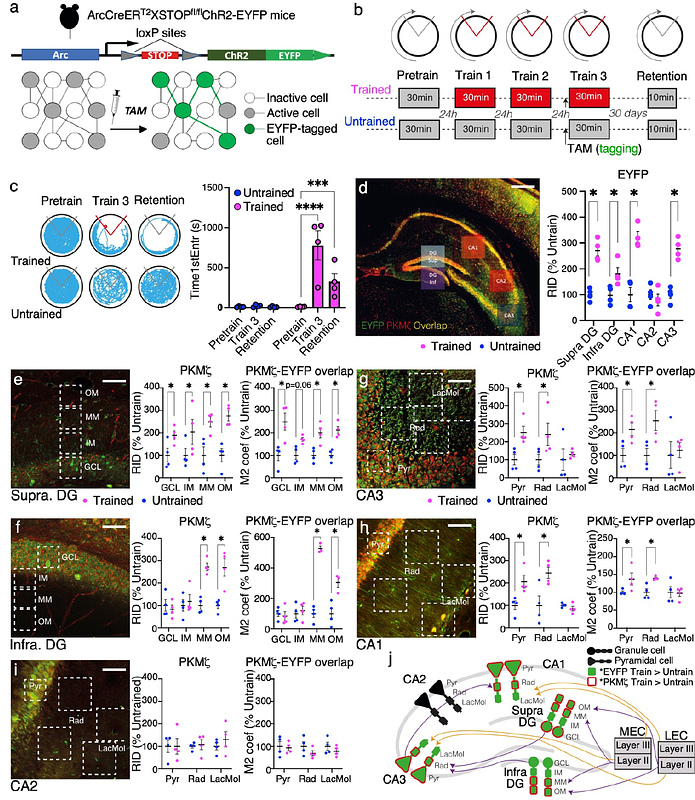
AbstractLong-term memory formation transiently activates Ca2+-calmodulin kinase II (CaMKII) and atypical protein kinase C isoform iota/lambda (PKC/{lambda}), whereas persistent activation of the other atypical PKC, protein kinase M zeta (PKM{zeta}), together with its interacting partner, the scaffolding-protein KIBRA, is necessary for maintaining potentiated synapses and memory. Here, we use immediate early gene (IEG) Arc activation during active place avoidance memory expression to tag memory-activated neurons with EYFP, followed by PKM{zeta} immunohistochemistry to learn which hippocampal synaptic pathways are persistently altered. For at least 1 month, PKM{zeta} and EYFP-PKM{zeta} colocalization persistently increase in the hippocampal tri-synaptic pathway (dentate gyrus (DG)[->]CA3[->]CA1). We thus use DG, CA3, and CA1 transcriptional profiling to identify mRNAs for PKM{zeta} and other molecules that might contribute to maintaining the memory. We find that memory persistence correlates with the upregulation of IEGs Arc, Fos, and NPas4 in DG. In contrast, expression of CaMKII, PKM{zeta}, PKC/{lambda}, KIBRA, and most other LTP-associated proteins do not covary with memory. This result rules out strong memory-related transcriptional regulation, but not regulation of mRNA translation or stability of these 'shadow proteins' that, despite being crucial for memory maintenance, evade detection by unbiased transcriptome profiling. We further examine whether covarying gene expression defines molecular ensembles that predict memory and Prkcz expression. Community detection, applied to both linear and non-linear pairwise gene co-expression relationships, reveals gene co-expression networks predictive of memory, memory-related IEGs, and Prkcz expression. These findings trace: i) a PKM{zeta} engram in the hippocampus; and highlight: ii) sustained IEGs upregulation in >24-h memory maintenance; iii) the utility of investigating both linear and non-linear gene expression covariance patterns; iv) limits of transcriptional profiling for identifying long-term memory molecules, most of which appear to be in 'shadow' ; and demonstrates v) that weak pair-wise gene-gene correlations can identify strong gene co-expression networks related to behavioral and shadow molecular estimates of long-term memory.