Parameter Optimization of Biochemical Models for Precision Medicine: A Case Study in PI(4,5)P2 Synthesis
Parameter Optimization of Biochemical Models for Precision Medicine: A Case Study in PI(4,5)P2 Synthesis
Hernandez-Hernandez, G.; Tieu, M.; Yang, P.-C.; Santana, L. F.; Clancy, C. E.
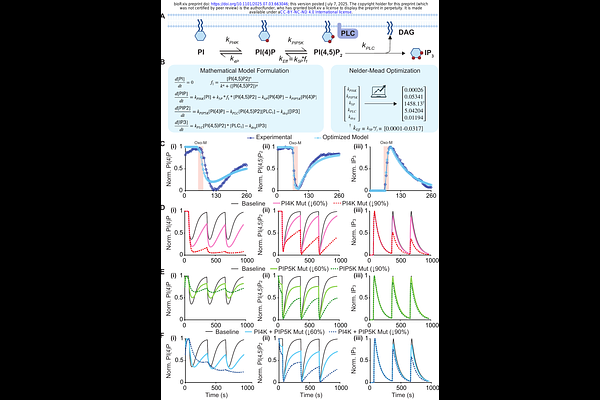
AbstractLipidomics is emerging as a domain of opportunity for precision medicine, with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) functioning as a central regulator of membrane signaling and metabolic control. Its rapid turnover and involvement in diseases like cancer and neurodegeneration highlight its dual role as a biomarker and therapeutic target. However, the inherent complexity of lipid networks complicates traditional modeling approaches. We present a modular, extendable framework for constructing mechanistic models of lipid kinetics via a case study, modeling the synthesis and degradation of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), a second messenger central to receptor signaling and calcium regulation. Our method efficiently identifies rate constants governing enzymatic dynamics and can be extended to a range of signaling networks. Using experimental time-course data for PI(4)P, PI(4,5)P2, and IP, we optimized five kinetic parameters to capture the lipid and second messenger dynamics. The resulting model achieved a strong correlation with experimental trends and reproduced dynamic behaviors relevant to cellular signaling. We then applied the model to simulate signaling perturbations linked to PI4KA and PIP5K1C loss-of-function, two lipid kinases associated with neurodevelopmental and neuromuscular disorders. This modeling framework provides a scalable foundation for predictive biochemical modeling and offers a path toward individualized applications in precision medicine.