A generative language model decodes contextual constraints on codon choice for mRNA design
A generative language model decodes contextual constraints on codon choice for mRNA design
Faizi, M.; Sakharova, H.; Lareau, L. F.
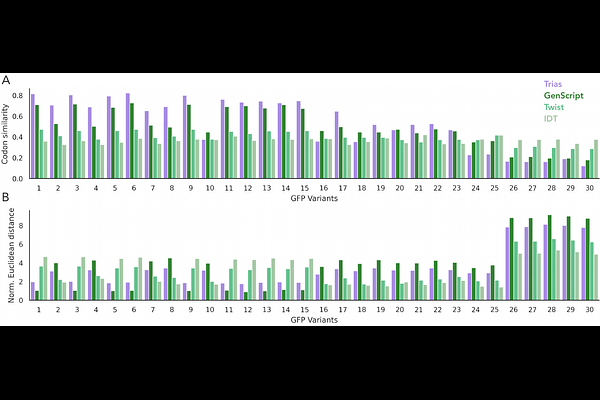
AbstractThe genetic code allows multiple synonymous codons to encode the same amino acid, creating a vast sequence space for protein-coding regions. Codon choice can have dramatic implications on mRNA function and protein output, and the impact of these choices has become newly relevant with advances in mRNA technology for vaccines and therapeutics. Genomes have evolved to prefer some codons over others, but simple methods for codon optimization that select the most preferred codons fail to capture complex contextual patterns in natural sequences. To enable biologically informed design of synthetic coding sequences, we present Trias, an encoder-decoder language model trained on the coding regions of millions of eukaryotic mRNA sequences. Trias learns codon usage rules directly from sequence data, integrating local and global dependencies to generate species-specific codon sequences that align with biological constraints. With no explicit training on protein expression, Trias can generate sequences and compute scores that correlate strongly with experimental measurements of mRNA stability, ribosome load, and protein output. The model outperforms commercial codon optimization tools in generating sequences resembling high-expression codon sequence variants. By capturing natural constraints on codon usage within sequence context, Trias provides a powerful, data-driven framework for synthetic mRNA design and for understanding the molecular and evolutionary principles governing codon choice.