Identification of functional genes in a chloroform and dichloromethane-degrading microbial culture
Identification of functional genes in a chloroform and dichloromethane-degrading microbial culture
Bulka, O.; Picott, K. J.; Mahadevan, R.; Edwards, E. A.
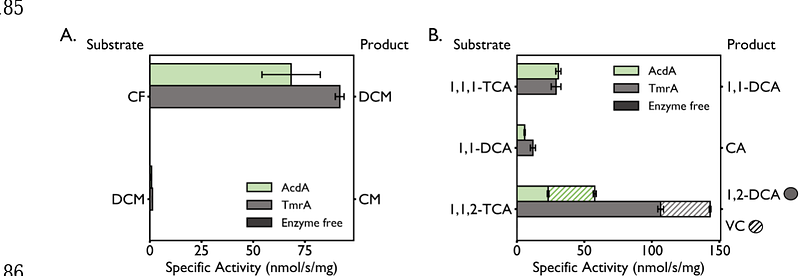
AbstractChloroform (CF) and dichloromethane (DCM) are groundwater contaminants of concern due to their high toxicity and inhibition of important biogeochemical processes. Biotransformation of CF and DCM have been well documented but always independent of one another. CF is dechlorinated to DCM by organohalide-respiring bacteria using reductive dehalogenases (RDases), while known DCM-degraders either ferment or mineralize DCM, both of which use the mec cassette to facilitate the entry of DCM to the Wood-Ljungdahl pathway. The SC05 culture, used commercially for bioaugmentation, is the first and only known stable enrichment culture to transform CF and DCM simultaneously. Here we use metagenomic and metaproteomic analysis to identify the functional genes involved in each of these transformations. A single Dehalobacter metagenome assembled genome contains the genes for an RDase, named acdA, and a complete mec cassette encoded on a single contig. AcdA and several of the Mec proteins were also highly expressed. Furthermore, AcdA displayed high dechlorination activity on CF and other chloroalkanes but did not show significant dechlorination of DCM. Overall, the high expression of Mec proteins and the activity of AcdA suggest a Dehalobacter capable of dechlorination of CF to DCM, and subsequent mineralization of DCM using the mec cassette.