Leveraging foundation models to dissect the genetic basis of cluster compactness and yield in grapevine
Leveraging foundation models to dissect the genetic basis of cluster compactness and yield in grapevine
Sharma, S.; Munoz, J.; Torres-Lomas, E.; Lin, J.; Banayad, H.; Lupo, Y.; Nunez, V.; Gaspar, A.; Cantu, D.; Diaz-Garcia, L.
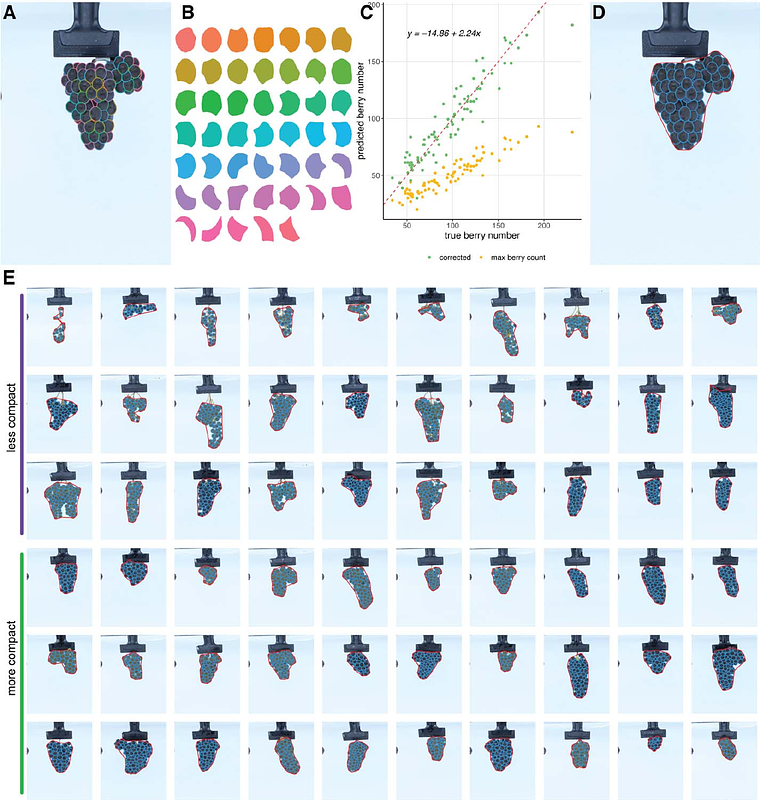
AbstractGrape cluster compactness is a key trait that influence fruit quality, yield, and disease susceptibility. Understanding the genetic basis of this trait is essential for optimizing vineyard management and improving grapevine cultivars. In this study, we performed quantitative trait locus (QTL) mapping to identify genomic regions associated with cluster architecture and yield components in a bi-parental population derived from Vitis vinifera cv. Riesling crossed to Cabernet Sauvignon. A total of 138 full-sibling progeny were evaluated over two growing seasons at Oakville, Napa Valley, California. Traditional yield-related traits were measured, including cluster number, total cluster weight, and average cluster weight. Additionally, an image-based phenotyping pipeline leveraging the foundation model Segment Anything Model (SAM) was employed to segment individual berries, measure their size and shape, and compute cluster compactness with minimal manual intervention. Trait correlations revealed that compact clusters tended to have a higher berry count but smaller berry size, highlighting the role of compactness in modulating cluster structure. Heritability estimates varied across traits, with berry dimensions and compactness displaying moderate to high heritability, indicating strong genetic control. Two parental linkage maps were constructed using a pseudo-test cross strategy. QTL mapping identified multiple loci associated with cluster architecture and yield components, with several stable QTLs detected across both years. Notably, a QTL for cluster compactness was found in both seasons on chromosome 1 in Cabernet Sauvignon. Other stable QTLs were associated with berry size (chromosomes 6 and 17) and berry count (chromosome 5 in Cabernet Sauvignon and chromosome 7 in Riesling). Additional QTLs were detected in a single year, reflecting the influence of environmental variation. Our findings provide valuable insights into the application of foundation models requiring no prior training and minimal intervention for high-quality segmentation and enhance our understanding of the genetic architecture of cluster compactness and yield traits. The genomic regions identified in this study offer promising targets for breeding programs aimed at improving grape quality and disease resistance.