Thermal proteome profiling identifies mitochondrial aminotransferases involved in cysteine catabolism via persulfides in plants
Thermal proteome profiling identifies mitochondrial aminotransferases involved in cysteine catabolism via persulfides in plants
Heinemann, B.; Moormann, J.; Bady, S.; Angermann, C.; Schrader, A.; Hildebrandt, T. M.
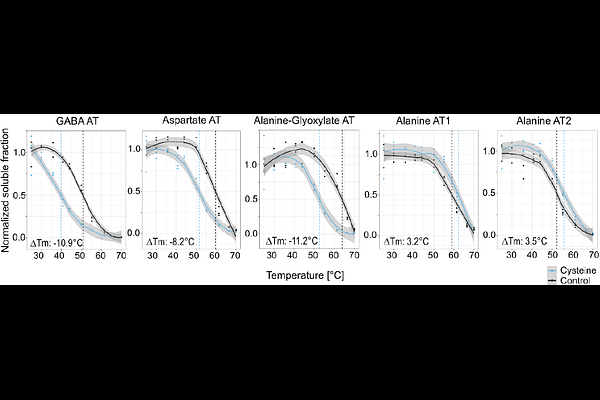
AbstractCysteine is a central metabolite in plant sulfur metabolism, with key roles in biosynthesis, redox regulation, and stress responses. While a mitochondrial cysteine degradation pathway has been described, the enzyme catalyzing its initial transamination step remained unidentified. Here, we applied thermal proteome profiling (TPP) to Arabidopsis mitochondria to uncover cysteine-interacting proteins. TPP successfully detected known cysteine-utilizing enzymes, validating its utility in plant metabolic research. Among newly identified targets were two aminotransferases annotated as alanine and aspartate aminotransferases that catalyze the transamination of cysteine to 3-mercaptopyruvate in vitro. These enzymes, together with the sulfurtransferase STR1 and the persulfide dioxygenase ETHE1, reconstituted a complete mitochondrial cysteine catabolic pathway. Kinetic data indicate that alanine aminotransferase, in particular, may function in vivo under physiological cysteine levels. Additionally, GABA aminotransferase was inhibited by cysteine, suggesting a regulatory role in stress metabolism. Beyond enzyme identification, the dataset provides a resource for exploring cysteine-mediated regulation of transporters, RNA-editing factors, and respiratory components. Given cysteine\'s emerging role as a metabolic signal in stress responses, and the importance of allosteric regulation in amino acid metabolism, these findings highlight the broader regulatory potential of cysteine-protein interactions in plants. This study demonstrates the utility of TPP for elucidating metabolite-protein networks and advancing our understanding of plant mitochondrial metabolism.