An optimised computational approach for the identification of somatic structural variants in cancer
An optimised computational approach for the identification of somatic structural variants in cancer
Waise, S.; Mensah, N.; Lesluyes, T.; Demeulemeester, J.; Flanagan, A. M.; Pillay, N.; Van Loo, P.
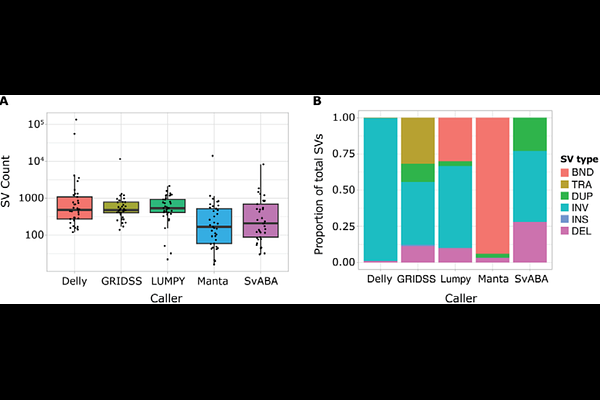
AbstractStructural variants play a critical role in tumorigenesis. At present, these events are most commonly identified using short-read whole-genome sequencing data, and a number of computational tools are available for this purpose. Consensus approaches have been used to improve precision, but may reduce sensitivity. The optimal number and combination of callers remains unclear, in part due to the lack of gold standard real-world datasets for validation. Here, we benchmark the performance of Delly, GRIDSS, LUMPY, Manta and SvABA, using a validation set of consensus calls from the Pan-Cancer Analysis of Whole Genomes Consortium. Manta showed the best standalone performance, identifying 88% of the validation set calls, and was included in all of the best-performing caller combinations. A consensus approach comprising Delly, GRIDSS, Manta and SvABA was selected as the optimum approach from those tested. We provide a NextFlow implementation of our optimised consensus approach as a resource for the cancer genomics community.