MonoAlg3D: Enabling Cardiac Electrophysiology Digital Twins with an Efficient Open Source Scalable Solver on GPU Clusters
MonoAlg3D: Enabling Cardiac Electrophysiology Digital Twins with an Efficient Open Source Scalable Solver on GPU Clusters
Berg, L. A.; Oliveira, R. S.; Camps, J.; Wang, Z. J.; Doste, R.; Bueno-Orovio, A.; dos Santos, R. W.; Rodriguez, B.
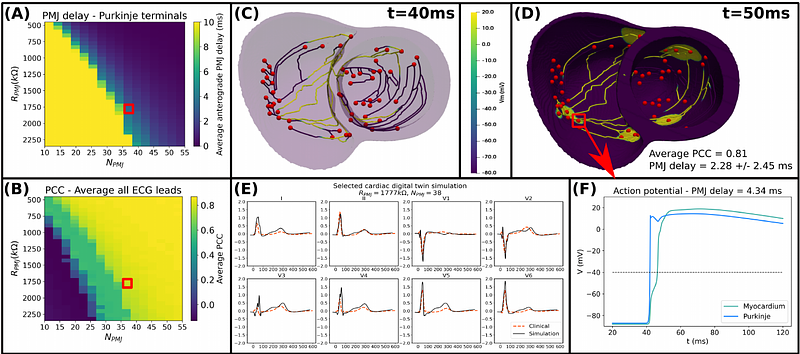
AbstractModelling and simulation are essential in biomedicine, and specifically in computational cardiology. Reliable, efficient and accurate solvers are critical. This study presents an open source, GPU-based cardiac electrophysiology solver for scalable digital twin multiscale simulations MonoAlg3D, incorporating conduction system calibration and performance optimization. The solver employs the monodomain equation coupled with the Purkinje network, solved via the finite volume method, featuring a GPU-based linear solver and concurrent simulation dispatch with MPI. We demonstrate a 10.94x speedup over a CPU-based solution and scalability by running 512 simulations on 128 compute nodes, completing all coarse-mesh simulations in less than 24 minutes and fine-mesh simulations in 303 minutes. We also demonstrate integration into a cardiac digital twin pipeline for personalisation based on clinical data. The proposed open source solver enhances computational efficiency and physiological fidelity, enabling large-scale, high-speed cardiac simulations. This work marks a significant step toward fast and scalable cardiac simulations on GPU architectures, with integration in a Digital Twin personalisation pipeline including the conduction system.