Monocyte single cell-type gene expression measured in peripheral blood by DIRECT LS-TA method: the ratio-based biomarkers of (IFI27/PSAP) showed superior performance than interferon score in triage patients with viral infection
Monocyte single cell-type gene expression measured in peripheral blood by DIRECT LS-TA method: the ratio-based biomarkers of (IFI27/PSAP) showed superior performance than interferon score in triage patients with viral infection
Tang, N. L.-s.; Kwan, T.-K.; Huang, D.; Huang, J.; Wang, X.; Lui, G. C.; Ma, S.-L.; Leung, K.-S.
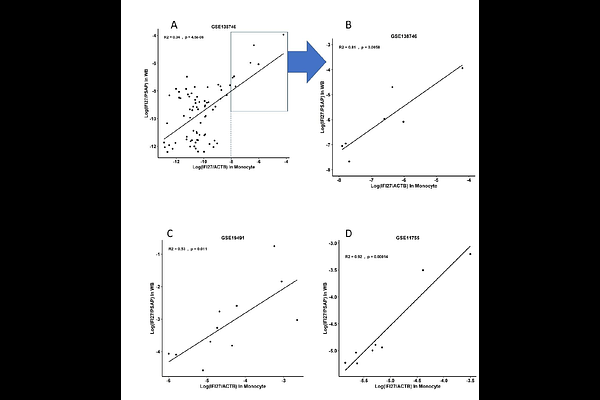
AbstractA rapid method to triage febrile patients into different categories of etiologies remains a significant challenge even nowadays, when many molecular tests for pathogens are available. Routine serum protein tests like C-reactive protein and procalcitonin have limited specificity. Host response gene signatures are promising biomarkers but they usually require assaying many genes, e.g. 7 genes are commonly used to calculate the interferon (IFN) score. However, these gene panels fail to capture cell-type-specific host responses. Measuring gene expression of a specified single cell population, like monocytes, offers enhanced biological insight. However, it currently requires laborious cell sorting or costly single-cell sequencing techniques, limiting its clinical applicability. This study aims to develop a simple ratio-based biomarker (RBB) representing monocyte-specific host response to viral infection called DIRECT LS-TA method. A simple ratio of 2 genes (both are shortlist monocyte informative genes) quantified in peripheral blood (PB) samples correlated with gene expression in purified monocytes in the corresponding individual. These RBBs cover 3 interferon-stimulated genes (ISGs): IFI27/PSAP, IFI44L/PSAP and SIGLEC1/PSAP. They are compared to the conventional multi-gene IFN score in the differentiation of viral infection. Public gene expression datasets from NCBI GEO were used to shortlist monocyte-informative genes that can be used as the RBB in PB. The DIRECT LS-TA RBB was calculated as the ratio of the target ISG transcript abundance (TA) to that of another reference gene (PSAP or CTSS) directly quantified from bulk PB data (e.g., Log(IFI27/PSAP) in WB). The correlation (expressed by coefficient of determination, R2) between these DIRECT LS-TA RBBs and the gold-standard target gene TA measured in purified monocytes was assessed. The diagnostic performance of selected RBBs (IFI27/PSAP, IFI44L/PSAP, SIGLEC1/PSAP) was compared against the conventional 8-gene IFN score for differentiating viral infections from controls. Direct LS-TA RBBs measured in PB showed strong correlation with gold-standard gene expression measured in purified monocytes (R2 ranged from 0.53 for the target gene IFI27 to >0.9 for the target gene IFI44L). This high level of correlation supports that this simple RBB (DIRECT LS-TA) method can replace the tedious cell sorting approach to obtain single-cell-type gene expression data. All DIRECT LS-TA results of ISGs were raised during viral infection. The best clinical performance in triaging viral infection patients was achieved by IFI27/PSAP or IFI27/CTSS across all datasets. For example, in the GSE111368 dataset, IFI27/PSAP achieved an AUC of 0.94 (95% CI 0.90-0.97) with 88% sensitivity and 95% specificity, surpassing the IFN score\'s AUC of 0.90 (95% CI 0.85-0.94) with 79% sensitivity and 93% specificity. Conclusion: The DIRECT LS-TA method, utilizing the format of simple two-gene ratio-based biomarkers like IFI27/PSAP, provides a robust and accurate measure of monocyte-specific interferon pathway activation directly from peripheral blood. The superior performance of the DIRECT LS-TA method makes it a promising, readily implementable tool for clinical triage. Its ability to provide single-cell-type specific information, rapid turnaround using standard qPCR/dPCR technology, and enhanced biological specificity make it a valuable molecular host response assessment.