Single-cell imaging reveals spontaneous phenotypic sorting and bet-hedging in developing biofilms
Single-cell imaging reveals spontaneous phenotypic sorting and bet-hedging in developing biofilms
Tai, J.-S. B.; Nam, K.-M.; Li, C.; Nijjer, J.; Zhang, S.; Waters, C. M.; Yan, J.
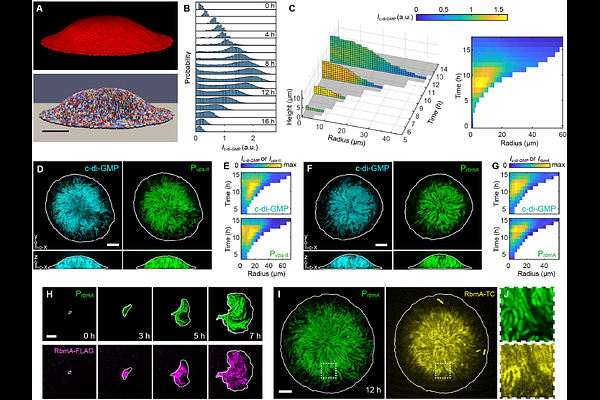
AbstractHow phenotypic heterogeneity shapes biofilm architecture and development remains poorly understood. Motivated by this, we developed imaging tools to track intracellular levels of c-di-GMP, a key second messenger that controls the motile-to-sessile transition, in developing Vibrio cholerae biofilms at single-cell resolution. We show that c-di-GMP levels spontaneously bifurcate into a bimodal distribution that forms a spatially sorted pattern: High-c-di-GMP cells dominate the biofilm core while low-c-di-GMP cells localize to the periphery. Combining single-lineage tracing, mutant analysis, and agent-based modeling, we reveal that this pattern arises from differential viscosity and surface friction mediated by matrix-dependent interactions between cells and their microenvironments. We demonstrate that this heterogeneity and phenotypic sorting enable continuous emergence and shedding of planktonic cells, enhancing fitness in fluctuating environments. Our findings uncover a differential drag mechanism for pattern formation in multicellular systems, and expand the classical picture of biofilm lifecycle by highlighting the functional significance of phenotypic heterogeneity.