Meta-Analysis of Urinary Metabolite GWAS Studies IdentifiesThree Novel Genome-Wide Significant Loci
Meta-Analysis of Urinary Metabolite GWAS Studies IdentifiesThree Novel Genome-Wide Significant Loci
Zaki, J. K.; Tomasik, J.; McCune, J. A.; Scherman, O. A.; Bahn, S.
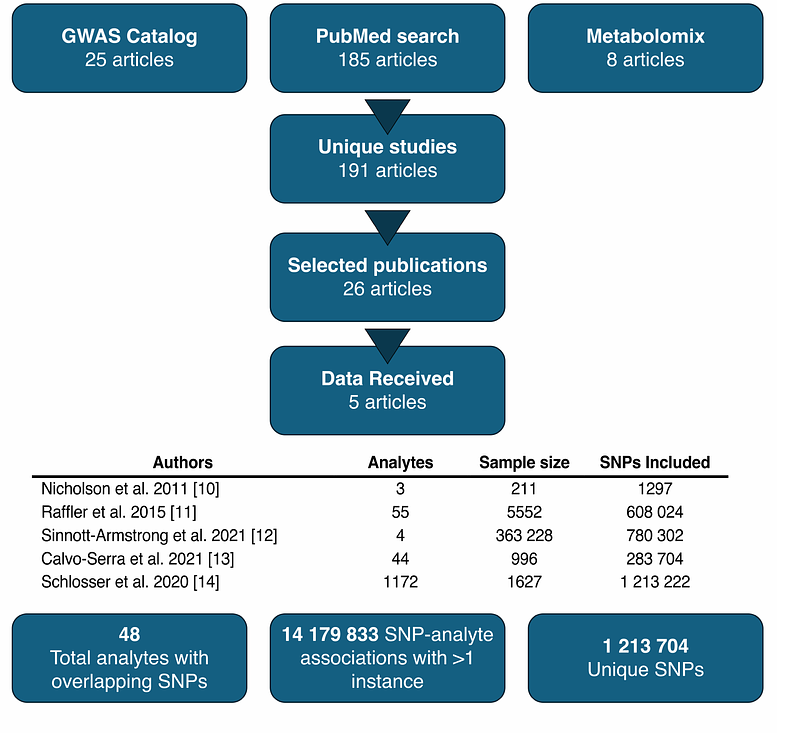
AbstractGenome-wide association studies (GWAS) have substantially enhanced the understanding of genetic influences on phenotypic outcomes; however, realizing their full potential requires an aggregate analysis of numerous studies. Here we represent the first comprehensive meta-analysis of urinary metabolite GWAS studies, aiming to consolidate existing data on metabolite-SNP associations, evaluate consistency across studies, and unravel novel genetic links. Following an extensive literature review and data collection through the EMBL-EBI GWAS Catalog, PubMed, and metabolomix.com, we employed a sample size-based meta-analytic approach to evaluate the significance of previously reported GWAS associations. Our results showed that 1226 SNPs are significantly associated with urinary metabolite levels, including 48 lead SNPs correlated with 14 analytes: alanine, 3-aminoisobutyrate, betaine, creatine, creatinine, formate, glycine, glycolate, histidine, 2-hydroxybutyrate, lysine, threonine, trimethylamine, and tyrosine. Notably, the results revealed three previously unknown associations: rs4594899 with tyrosine (P = 6.6 x 10-9, N = 5447), rs1755609 with glycine (P = 3.3 x 10-10, N = 5447), and rs79053399 with 3-aminoisobutyrate (P = 6.9 x 10-10, N = 4656). These findings underscore the potential of urinary metabolite GWAS meta-analyses in revealing novel genetic factors that may aid in the understanding of disease processes, and highlight the necessity for larger and more comprehensive future studies.


