A single-cell multiomics roadmap of zebrafish spermatogenesis reveals regulatory principles of male germline formation
A single-cell multiomics roadmap of zebrafish spermatogenesis reveals regulatory principles of male germline formation
Ruiz, A. M. B.; Geng, F.-S.; Pujol, G.; Sanabria, E.; Brethouwer, T.; Castillo, M. A.; Ruiz-Herrera, A.; Tena, J. J.; Bogdanovic, O.
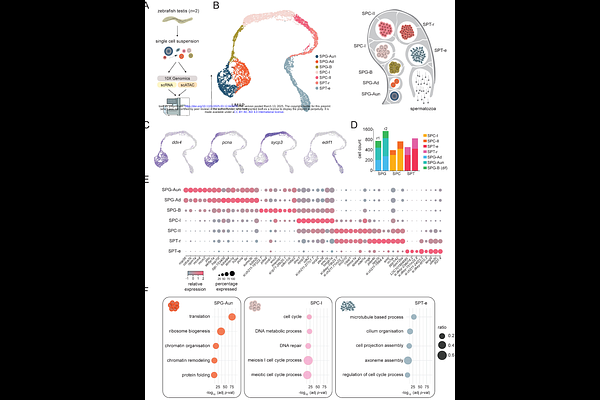
AbstractSpermatogenesis is the biological process by which male sperm cells (spermatozoa) are produced in the testes. Beyond facilitating the transmission of genetic information, spermatogenesis also provides a potential framework for inter- and transgenerational inheritance of gene-regulatory states. While extensively studied in mammals, our understanding of spermatogenesis in anamniotes remains limited. Here we present a comprehensive single-cell multiomics resource, combining single-cell RNA sequencing (scRNA-seq) and single-cell chromatin accessibility (scATAC-seq) profiling, with base-resolution DNA methylome (WGBS) analysis of sorted germ cell populations from zebrafish (Danio rerio) testes. We identify major germ cell types involved in zebrafish spermatogenesis as well as key drivers associated with these transcriptional states. Moreover, we describe localised DNA methylation changes associated with spermatocyte populations, as well as local and global changes in chromatin accessibility leading to chromatin compaction in spermatids. Notably, we identify loci that evade global chromatin compaction, and which remain accessible, suggesting a potential mechanism for the intergenerational transmission of gene-regulatory states. Overall, this high-resolution atlas of zebrafish spermatogenesis provides a valuable resource for studying vertebrate germ cell development, evolution, and epigenetic inheritance.