ntSynt-viz: Visualizing synteny patterns across multiple genomes
ntSynt-viz: Visualizing synteny patterns across multiple genomes
Coombe, L.; Warren, R. L.; Birol, I.
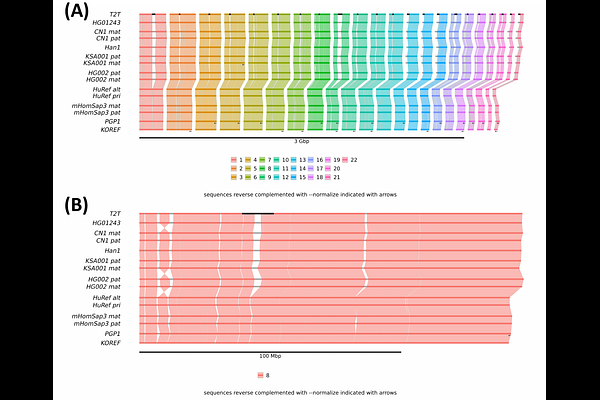
AbstractWith the explosion of chromosome-scale genome assemblies being generated in recent years, there is vast potential for comparative genomics analyses through detecting multi-genome synteny. While existing tools can detect synteny blocks between multiple genomes, their text-based outputs make it challenging to intuitively explore large-scale synteny patterns. Interpretable, information-rich and easy-to-use synteny visualization tools are imperative to enable important biological insights from the synteny block data output by the aforementioned utilities. Here, we present ntSynt-viz, a command-line tool for automated sorting, normalization and plotting of multi-genome synteny blocks. We show how ntSynt-viz provides clearer and more easily interpretable chromosome-painting ribbon plots compared to the state-of-the-art tool NGenomeSyn when evaluating synteny between 14 human genomes and 9 hoverfly genomes. We expect that ntSynt-viz will provide crucial insights into large-scale synteny patterns between divergent genomes, thereby advancing research into key evolutionary questions. ntSynt-viz is freely available on GitHub (https://github.com/bcgsc/ntsynt-viz).