Loss of function of chromatin remodeler OsCLSY4 leads to RdDM-mediated mis-expression of endosperm-specific genes affecting grain qualities
Loss of function of chromatin remodeler OsCLSY4 leads to RdDM-mediated mis-expression of endosperm-specific genes affecting grain qualities
Pal, A.; Rana, S.; Dey, R.; Shivaprasad, P. V.
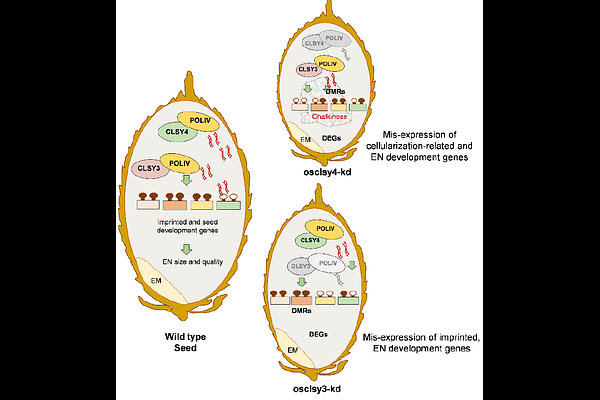
AbstractRNA-directed DNA methylation (RdDM) sequence-specifically targets transposable elements (TEs) and repeats in plants, often in a tissue-specific manner. In triploid endosperm tissue, RdDM also acts as a parental dosage regulator, mediating spatio-temporal expression of genes required for its development. It is unclear how RdDM is initiated and established in endosperm. Rice endosperm-specific imprinted chromatin remodeler OsCLSY3 recruits RNA polymerase IV to specific genomic sites for silencing and optimal gene expression. Here we show that, in addition to OsCLSY3, ubiquitously expressed OsCLSY4 is also crucial for proper reproductive growth and endosperm development. Loss of function of OsCLSY4 led to reproductive and nutrient-filling defects in endosperm. Using genetic and molecular analysis, we show that OsCLSY3 and OsCLSY4 play both overlapping and unique silencing roles in rice endosperm by targeting specific and common TEs, repeats and genic regions. These results indicate the importance of optimal expression of two OsCLSYs in regulating endosperm-specific gene expression, genomic imprinting and suppression of specific TEs. Results presented here provide new insights into the functions of rice CLSYs as upstream RdDM regulators in rice endosperm development, and we propose that functions of their homologs might be conserved across monocots.