DNA six-way junction conformations and their use in a 2D square lattice
DNA six-way junction conformations and their use in a 2D square lattice
Baten, A. J.; Mason, H. G.; Veneziano, R.; Bricker, W. P.
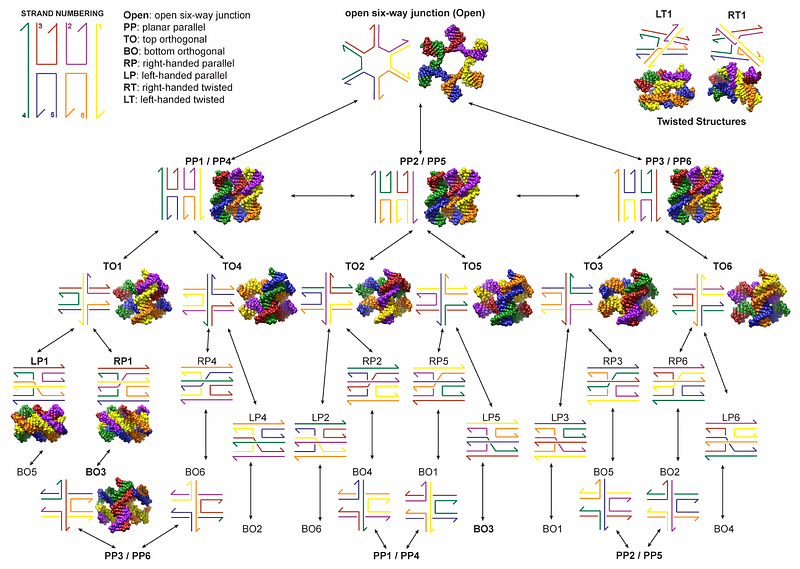
AbstractIn this paper, we investigate the conformational landscape of the DNA six-way junction (6WJ), a higher-order extension of the DNA four-way junction (4WJ), to determine preferred structural isomers. The 6WJ allows for unique junction-level topologies, and could be used to create novel DNA nanostructures, but the conformational landscape for the 6WJ is much more complex than the 4WJ. Our proposed conformational landscape for the 6WJ includes eight unique structural motifs, including an unstacked motif, five distinct stacked motifs, and two twisted motifs, and we estimate that there are forty structural isomers for the 6WJ, compared to only three for the 4WJ. To gain insight into these conformations, we perform all-atom molecular dynamics (MD) simulations on fourteen of the structural isomers. Our analysis shows that each 6WJ motif can be distinguished by its duplex stacking angles, and only a few of the 6WJ isomers have favorable free energies, which include the planar parallel isomers, the twisted isomers, and one of the orthogonal isomers. Lastly, to confirm that these stacked 6WJ motifs could be used to create larger-scale DNA nanostructures, we design a 2D square lattice using four orthogonal motifs connected in a 2x2 arrangement. Experimental characterization of this 2D lattice shows folding into the correct size and shape, further confirmed by AFM images of the DNA tiles, and an MD simulation that shows a slight twist of the lattice structure from orthogonality. This is the first 2D tile assembly built using an orthogonal junction-level topology for DNA nanotechnology, allowing a smaller mesh size than is possible with the 4WJ architecture.