Epigenome and interactome profiling uncovers principles of distal regulation in the barley genome
Epigenome and interactome profiling uncovers principles of distal regulation in the barley genome
Navratilova, P.; Pavlu, S.; Zhu, Z.; Tulpova, Z.; Kopecky, O.; Novak, P.; Stein, N.; Simkova, H.
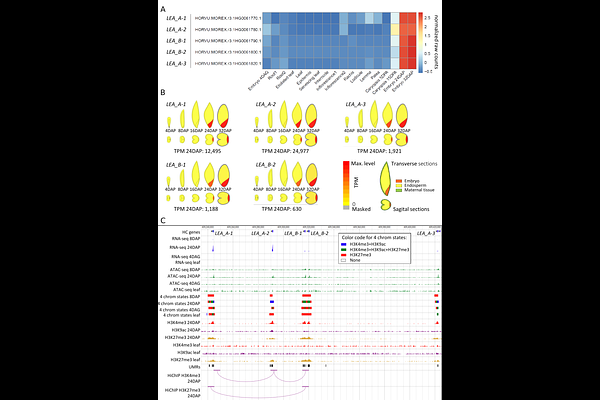
AbstractRegulation of transcription initiation is the ground level of modulating gene expression during plant development. This process relies on interactions between transcription factors and cis-regulatory elements (CREs), which become promising targets for crop bioengineering. To annotate CREs in the barley genome and understand mechanisms of distal regulation, we profiled several epigenetic features across three stages of barley embryo and leaves, and performed HiChIP to identify activating and repressive genomic interactions. Using machine learning, we integrated the data into seven chromatin states, predicting ~77,000 CRE candidates, collectively representing 1.43% of the barley genome. Identified genomic interactions, often spanning multiple genes, linked thousands of CREs with their targets and revealed notably frequent promoter-promoter contacts. Using the LEA gene family as an example, we discuss possible roles of these interactions in transcription regulation. On the Vrn3 gene, we demonstrate the potential of our datasets to predict CREs for other developmental stages.