Sequencing and functional analysis of Sphingobium yanoikuyae A-TP genome reveals genes for utilization of limonene, α-pinene, and citronellol
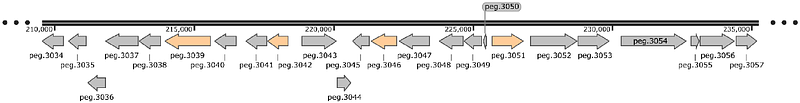
Sequencing and functional analysis of Sphingobium yanoikuyae A-TP genome reveals genes for utilization of limonene, α-pinene, and citronellol
Jankovic, N.; Marques de Castro, G.; Quintanilha Torres, A. L.; Chelala Moreira, R.; Uliano-Silva, M.; Souza da Conceicao, C.; Lima Coelho, V.; Alves Americo, J.; Lemos Bicas, J.; de Freitas Rebelo, M.
AbstractSphingobium yanoikuyae A-TP is a bacterial strain capable of utilizing and biotransforming commercially relevant monoterpenes. Genome mining and similarity-based analysis identified two gene clusters (24 and 30 genes) likely responsible for limonene, -pinene, and citronellol metabolism. Notably, genes associated with -pinene metabolism - an oxide lyase and an aldehyde dehydrogenase - were clustered with those for citronellol utilization, despite limonene closer structural similarity to -pinene. Growth experiments confirmed citronellol as a carbon source, but linalool, geraniol and {beta}-myrcene were not substrates for this strain. To address the unresolved identity of limonene 8-hydratase, the enzyme catalyzing limonene conversion to -terpineol, we performed RNA-seq analysis on strains with contrasting phenotypes which identified highly upregulated candidate genes. Phylogenetic analysis supports the classification of this strain as a Sphingobium yanoikuyae species. This study advances the understanding of monoterpene catabolism in Sphingomonadaceae and provides a foundation for biotechnological production of monoterpenes and their derivatives.